Bioimpacts. 2025;15:29944.
doi: 10.34172/bi.29944
Review
Microfluidics as a promising technology for personalized medicine
Zahra Oushyani Roudsari Investigation, Visualization, Writing – original draft, Writing – review & editing, 1, 2 
Zahra Esmaeili Investigation, Writing – original draft, 3
Nafiseh Nasirzadeh Investigation, Writing – original draft, 3
Saeed Heidari Keshel Validation, 4, 5
Farshid Sefat Data curation, Formal analysis, Validation, 6, 7
Hassan Bakhtyari Validation, 8
Samad Nadri Conceptualization, Data curation, Formal analysis, Methodology, Project administration, Supervision, Validation, 9, 10, 3, * 
Author information:
1Student Research Committee, School of Medicine, Zanjan University of Medical Sciences, Zanjan, Iran
2Department of Medical Biotechnology, School of Medicine, Zanjan University of Medical Sciences, Zanjan, Iran
3Department of Medical Nanotechnology, School of Medicine, Zanjan University of Medical Sciences, Zanjan, Iran
4Medical Nanotechnology Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
5Department of Tissue Engineering and Applied Cell Science, School of Advanced Technologies in Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
6Department of Biomedical and Electronics Engineering, School of Engineering, University of Bradford, Bradford, UK
7Research Centre in Polymer Science & Technology (Polymer IRC), University of Bradford, Bradford, UK
8Department of Pediatrics, School of Medicine, Ayatollah Mousavi Hospital, Zanjan University of Medical Sciences, Zanjan, Iran
9Zanjan Pharmaceutical Nanotechnology Research Center, Zanjan University of Medical Sciences, Zanjan, Iran
10Zanjan Metabolic Diseases Research Center, Health and Metabolic Diseases Research Institute, Zanjan University of Medical Sciences, Zanjan, Iran
Abstract
Introduction:
Due to the recent advances in biomedicine and the increasing understanding of the molecular mechanism of diseases, healthcare approaches have tended towards preventive and personalized medicine. Consequently, in recent decades, the utilization of interdisciplinary technologies such as microfluidic systems had a significant increase to provide more accurate high throughput diagnostic/therapeutic methods.
Methods:
In this article, we will review a summary of innovations in microfluidic technologies toward improving personalized biomolecular diagnostics, drug screening, and therapeutic strategies.
Results:
Microfluidic systems by providing a controllable space for fluid flow, three-dimensional growth of cells, and miniaturization of molecular experiments are useful tools in the field of personalization of health and treatment. These conditions have enabled the potential to carry out studies like; disease modeling, drug screening, and improving the accuracy of diagnostic methods.
Conclusion:
Microfluidic devices have become promising point-of-care (POC) and personalized medicine instruments due to their ability to perform diagnostic tests with small sample volumes, cost reduction, high resolution, and automation.
Keywords: Microfluidic systems, Personalized medicine, Biomolecular diagnostics, Drug screening, Therapeutic strategies
Copyright and License Information
© 2025 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Funding Statement
None.
Introduction
During the recent decades, microfluidic technology has become widely used in biology research. Microfluidics is an interdisciplinary science in which various disciplines such as biotechnology, engineering, and physics are involved. The microfluidic device is made of specific structures—including very small-scale channels—that enable the manipulation of small volume of fluids at microliter or picolitre levels 1 (Fig. 1).
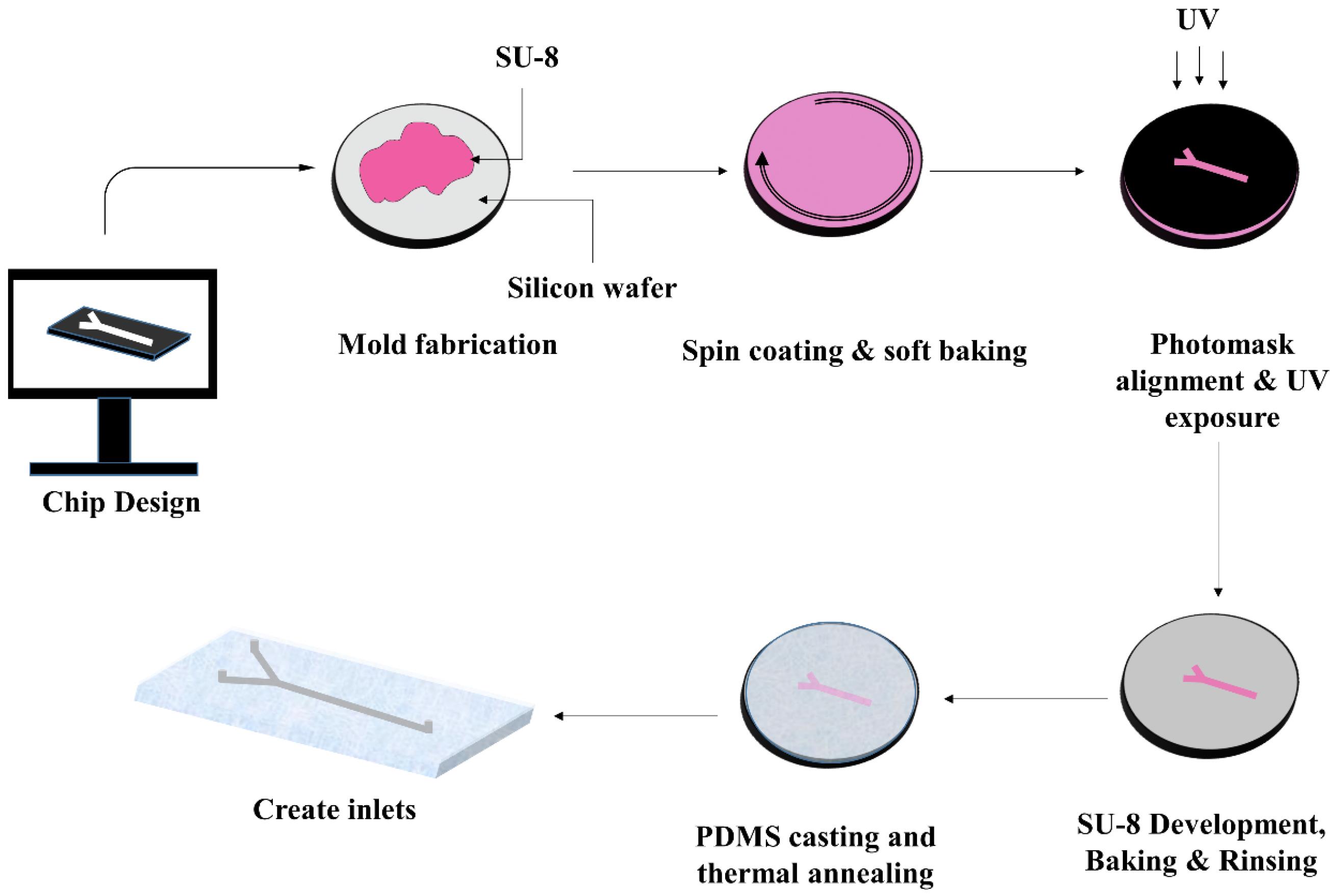
Fig. 1.
Schematic steps of fabricating a typical PDMS microfluidic device via soft lithography method.
.
Schematic steps of fabricating a typical PDMS microfluidic device via soft lithography method.
These attributes have several advantages: a remarkable decline in the use of resources (such as expensive/tracer reagents or samples), a more accelerated experimental process, extra precise control over the experiment. These features make this technology an ideal tool for using in biological researches. Another important feature of microfluidics is that laminar flow assists the simulation of interstitial flows within the body, the spatial orientation of cells, and the gradient of concentration of various chemical factors.2 Hence, microfluidic technology enables the culture and study cell behavior in a controlled condition.3 Surface modification and coating of the polydimethylsiloxane (PDMS) and glass, as a mostly applied materials for the fabrication of microfluidic devices,4 by extracellular matrix (ECM) components such as fibronectin, collagen, or matrigel improves cell attachment, and then cell proliferation.5,6
Conventionally, research on human diseases and drug screening was performed on 2D cell culture using petri dishes that is oversimplified and not possible to mimic microenvironment complexity and physiological condition in living tissues. Therefore, results obtained from 2D models may not be completely reliable.7,8 On the other hand, even though physiologic environments can be found in animal-based systems, but the use of animal models is sometimes problematic, and the results in this system are not always generalizable to humans. These hitches eventually led to the development of 3D cell culture based approaches that provide both the physiologic microenvironment of the cells and the ability to more accurately control the cell conditions.9
Nowadays, microfluidic devices providing a 3D platform for cell culture enabled the reconstruction of tissues and organs. In this regard, numerous microfluidic devices with complex microstructures and substantial biochemical/bioelectrical performance have been achieved. These microfluidics platforms have the potential to be extended for 3D biomimetic tissues and organotypic culture for a wide range of purposes.10-13 Therefore, in this article, first, explanations about personalized medicine and microfluidic systems were presented. Then we discussed the application of microfluidic systems that have been recently developed in various fields of biomedicine, including molecular diagnosis, oncology studies, drug screening, and organ-on-a-chip with a personalized medicine approach.
Personalized medicine and microfluidic technologies
Over the past decades, increasing knowledge on the molecular mechanism of gene expression and mutation has revealed the mechanism of many diseases, and researchers have used this information to bridge the gap between experimental researches and therapeutic approaches.14 In this regard, scientists are applying genetic-based diagnostic tests and molecular mechanisms to more accurately predict patients' response to targeted therapies. Therefore, nowadays most efforts have focused on developing new treatments and optimizing prescriptions by directing patients to the best drug, appropriate dose at the right time.15
Completion of the human genome project in 2003 opened new horizons for medical science. The data from this project has helped a great deal in medical science, especially in the field of oncology.16,17 as it provided valuable information on gene sequences, polymorphisms and mutations. Presented genetic data have important applications such as prognostic value and significant therapeutic implications.18 The analysis of these data led us to realize that genetic differences in individuals will affect almost all aspects of the physiological function of individuals. In fact, the human genome project offered new perspectives to a better understanding of the genetic origins of individual differences and their application in the medical fields.18,19 This developing field that uses risk factors, molecular diagnostics, targeted therapies, and pharmacogenomics to improve the efficacy of treatment is called personalized medicine. Personalized medicine links one's molecular signature to its medical profile to help to make the right decisions about the patient's treatment.15,20
Studying the molecular markers (DNA, protein, or mRNA) has changed our view of diseases and also created molecular classification for diseases. Genetic variations in individuals such as polymorphisms and even environmental effects can influence the uptake, excretion, distribution, and metabolism of drugs (for instance through changes in enzyme kinetics), thereby affecting the response to treatment.21 Therefore, approaches based on the origin of the disease will replace conventional therapies. In this regard, a pharmacogenomic test will be needed to evaluate the patient's therapeutic response to the drug based on the patient's genomic profile. It can be expected that; personalized medicine can make a big breakthrough in the pharmaceutical industry and medical practices in the not-too-distant future.22,23 Alterations in the medical system's pattern from reactive treatment to early detection or pro-active prevention, reduction of trial-and-error in the treatments and formation of an information-based treatment system for patient management are some of expected changes.24
Despite advances in the molecular genetics and medical fields, there are still some obstacles in providing appropriate genetic analysis methods or disease models. Hence, it is vital to produce models that are compatible with each patient's genetic system for clinical study. Personalized disease model organoids can be a powerful tool in the determination of treatment approaches. These organoids can be used as live models for investigating the effects of drugs, gene editing, surgery or even preventive studies.25,26 In this regard, various studies have demonstrated the potential of microfluidic technique to improve the similarity of 3D cell culture models to in-vivo physiological conditions and organoid construction.27,28
Microfluidic can be a precise and quick tool for clinical diagnostics. The microfluidic device is capable of using whole blood and giving a genetic profile endpoint.29 Microfluidic genetic analysis (MGA) systems can be applied for nanoliter flow control, electrophoresis, DNA purification and PCR amplification. Besides, these MGA systems have significant advantages such as reduced time and reagent consumption per trial, a reduced operator based variability or errors and providing better safety for the operator.30,31 Microfluidic systems can also be used to extract exosome from bodily fluids as a liquid biopsy for personalized medicine. Exosomes contain valuable information such as disease status, treatment response, exposure to environmental symptoms, and numerous other health factors that have diagnostic and therapeutic applications.32 Thus, eventually, we expect that microfluidic technology will create a leap in personalized medicine era.
Application of microfluidic in biomedicine
Microfluidic technology has become the ideal model for biomedical researches.33 In this article, practical examples of the microfluidic system in various aspects of biomedicine will be discussed as well as diagnosis, prognosis, and treatment with regards to the personalized medicine approach.
Biomolecular diagnostics
Over the past decades, biomolecule diagnostic methods have been widely used in medicine since can provide highly valuable information for detection, characterization, and prognosis of diseases. Foreign DNAs, modified gene expression levels, specific proteins, and genetic mutations are sometimes linked to pathologic conditions such as infectious diseases, various cancers and genetic disorders.34 Early detection, especially in the cases of high mortality diseases, infectious diseases, and cancer, has received much attention today. Early diagnosis considerably reduces the mortality rate and cost of treatment.35 On the other hand, personalizing a molecular diagnosis based on each person's molecular pattern can provide more accurate and precise diagnosis, optimizes treatment, and helps to implement an effective medication (pharmacogenomics). Immunoassay based methods such as ELISA or blotting techniques are routinely used in biomolecular diagnosis but, these methods typically require several steps, long incubations, and more consumption of reagents and samples. PCR has improved these drawbacks but there is still a need for faster, more sensitive and affordable techniques.36 Thus, developing methods for measuring faster, more accurately, more sensitively and in small quantities of the biomolecules have become a challenge in this field and various techniques have been proposed accordingly.37 The microfluidic and lab on chip strategies have become a critical tool in biomolecule diagnosis by providing these features (Table 1).38
Table 1.
Microfluidic-based studies for biomolecular detection
|
Target biomolecule
|
Microfluidic platform
|
Ref
|
| Rapid capture and fluorescent detection of rolling circle amplification products (RCPs) |
Combination of silica-based microfluidic device with thin film photodiodes to capture and detect florescent labeled rolling circle amplification (RCA) product |
39
|
| Isolation of circulating tumor cells (CTCs) from prostate cancer patients |
Functionalized microfluidic device with dual antibodies (anti-PCA and anti-EpCAM) |
40
|
| Tau Protein |
A fiber-optic sensor containing anti-tau antibodies and a SnO2−x thin film in which the binding of serum tau protein to anti-tau can be sensed via SnO2-mediated wavelength shifts. |
41
|
| Glucose |
A microfluidics containing microneedle patch for transdermal biofluid collection and an electrochemical biosensor for high sensitivity glucose detection |
42
|
| miR-21 |
Binding of miR-21 to the hairpin probe leads to the exposure of a single-strand fragment of the hairpin to the electrochemical probe and initiation of an electrochemical process |
43
|
| Detection and quantification of IL-6 and PSA proteins |
An ECL based immuno-detection microfluidic device with IL-6 and PSA capture Ab coated channels. Secondary Abs are coated on a silica nanoparticle |
44
|
| Detection of disease by analyzing of physical features of blood |
A microfluidic device consisted of capillary scanning viscometer and pendant drop tensiometer combined with system architecture for disease topography |
45
|
| miRNAs detection |
Power-free microfluidic chip for miRNAs detection using a sandwich hybridization system |
46
|
| The simultaneously detection of inflammatory biomarkers |
Combination of localized surface plasmon resonance (LSRP) sensor with one layer-four channel microfluidic device which utilized by automated microfluidic control |
47
|
The miniaturization of molecular techniques such as PCR on microfluidic devices will considerably reduce the sample loss, cost and laboratory space required for the processes as the entire procedure will be performed on a single chip. Moreover, simultaneous analysis of multiple samples with precision at the single cell or single gene level is also possible.48,49 Extracellular vesicles (EVs) are also valuable biomarkers, containing nucleic acids and proteins. investigation of EVs provides valuable data about the patient's condition. Hence, their confinement from biological liquids could be a potential diagnostic strategy. In this respect, microfluidic frameworks are utilized as a reasonable instrument for isolating EVs (Fig. 2).50
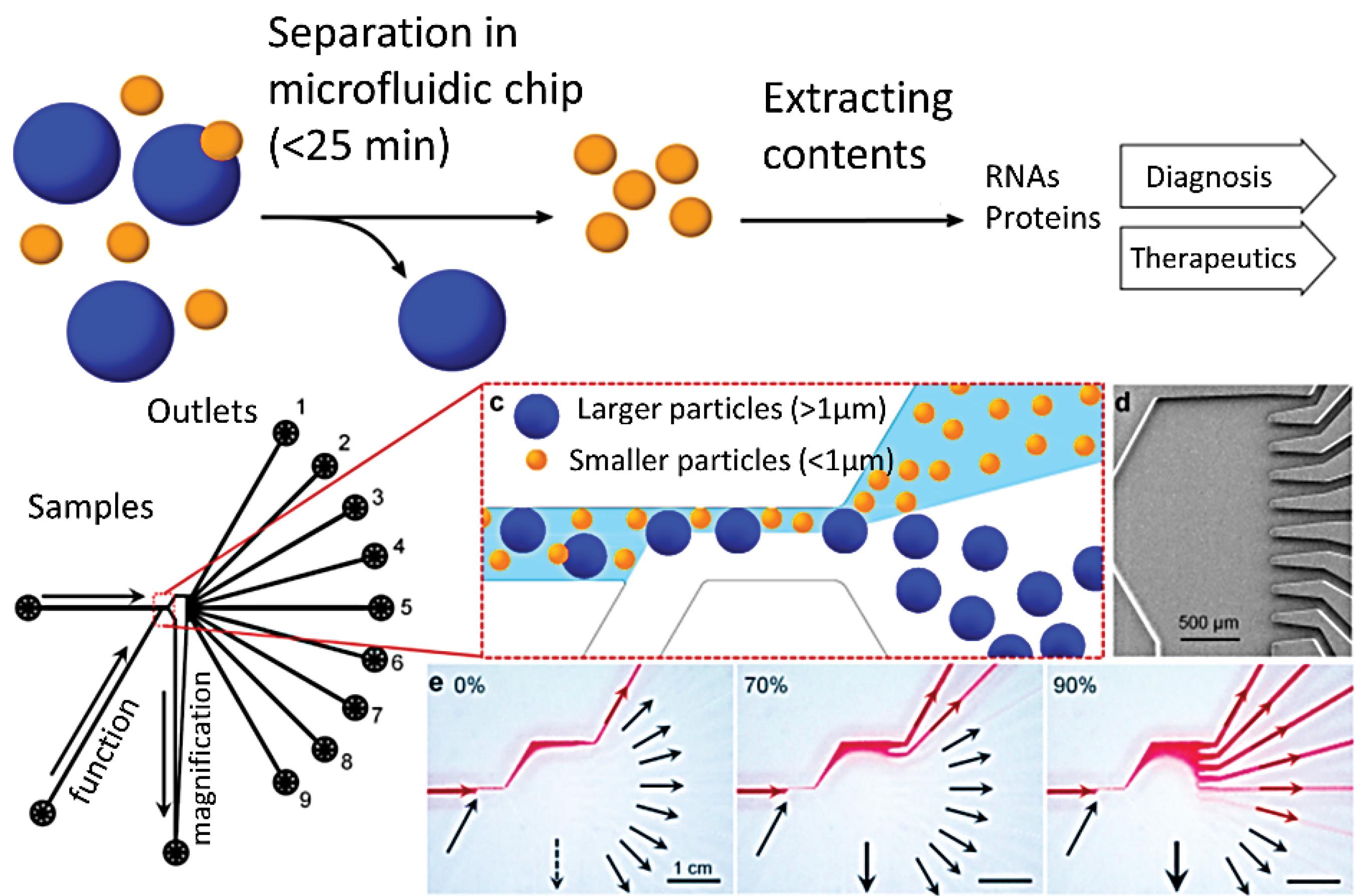
Fig. 2.
Atunable microfluidic devicefornoninvasive size-based separation of EVs. Reprinted from Shin et al50 under a Creative Commons Attribution 4.0 International.
.
Atunable microfluidic devicefornoninvasive size-based separation of EVs. Reprinted from Shin et al50 under a Creative Commons Attribution 4.0 International.
Several microfluidic platforms with the ability to nucleic acid extraction, purification, PCR amplification, electrophoresis, and DNA microarray have been developed lately.48,49 In this area, a nucleic acid purification chip device based on the use of silicon beads has been developed. In this device, the mixing of the beads with the sample is regulated through the buttons installed, by controlling the flow rate and the valves. A reciprocating flow is applied in this device in order to recover the sample in the system and increase the purification efficiency.51 In another study, a button actuation chip consists of two plasma separation and nucleic acid extraction/purification parts was designed to detect the nucleic acid of pathogenic bacteria in blood samples. Silica-coated magnetic nano particles (MNPs) is used to separate bacteria from plasma, and the steps of lysis and washing are carried out by the buttons installed on the device that control the pumps and valves.52 Integrated platforms are also designed based on a variety of electrophoretic methods such as capillary electrophoresis (CE),53 capillary gel electrophoresis (CGE),54 isoelectric focusing (IEF) and SDS-PAGE.55-57 Immuno-affinity miniaturization devices are also developed as another on-chip biomolecular diagnostic tools that have some advantages over conventional methods such as enhanced performance, speed up and simplify the process as well as employing lower volumes.58 On-chip immune-affinity approaches are based on detecting key biomarkers associated with the various diseases such as EGFR (head and neck cancer) and C-reactive protein (CRP) as a biomarker of coronary heart diseases,59 detecting vascular-endothelial growth factor (VEGF), prostate-specific antigen (PSA), and PCa circulating tumor cells (CTC) in human serum. For fabrication of this device first, aptamer and antibody have been coated on gold nanorods (GNR) and a UV-Vis-NIR spectrophotometer have been used as the detection system.60
Among the various instruments designed, Lee and colleagues developed a simple and rapid colorimetric system for the simultaneous detection of multiple microRNAs. MicroRNAs (miRNAs) are small (typically 18 to 24 bases) non-coding RNAs, which regulate gene expression by binding to 3´UTR of target mRNAs and inducing their degradation.61,62 The recent discoveries have suggested miRNAs as promising biomarkers for various diseases,63 drug sensitivity/resistance, and associating with personalized medicine64 and so, several microfluidic-based methods for miRNAs detection have been invented.65 Lee and colleagues have been used polyethylene glycol hydrogel as a substrate to facilitate the hybridization of microarrays with designed probes. Also, detection is done by colorimetric examination of gold nanoparticles attached to the target using dark field imaging.66 A droplet microfluidic high throughput device 300-500 cells/minute with analysis efficiency provided by Gou et al. This continuous-flow microfluidic platform utilizes two hairpin DNAs structures for capturing target miRNA from the lysed cell. Conformational changes in hairpin DNAs after binding lead to the separation of fluorophore and quencher pairs and eventually forming a signal senses by a UV detector.67 Additionally, microfluidic paper-based analytical devices (µPADs) are also designed as a biosensor for semi quantitative/quantitative detection goals. Sun, et al developed a paper-based electrochemical/visual biosensor for miRNA detection. In this platform, a biocompatible porous cellulose fiber web (µPADs) is modified with Au-NRs which assists the hairpin DNA probes to be immobilized on the surface by Au-S bonds. When the target miRNA binds to the hairpin probe, a single-strand fragment of the hairpin will be exposed and binds to the electrochemical probe (CeO2-Au@GOx).68
Different types of microfluidics-based biosensors are proposed such as glucose biosensors, urea biosensors, cholesterol biosensors, toxin detection biosensors.69 Biosensors are one of the most widely used molecular diagnostic tools due to their speed and ease of performance and hence their combination with microfluidic technology will improve their usage in molecular diagnostics. For instance, optofluidic devices can be categorized based on optical detection methods such as plasmon resonance technique, fluorescence, and interferometry. The integration of microfluidics with optical detection systems will reduce hardware costs and improve their sensitivity.70,71 The optofluidic devices are usually consist of three layers: The topmost layer consists of the microfluidic controls such as valves and pumps, the middle layer with microfluidic channels, and the third layer that contains the optical structures, sensors, and waveguides.72 A metal film or fluids can be used for coupling optical functionalities on-chip.73 One of the promising platforms of the optofluidic biosensor is a chip designed based on a liquid crystal system. In this device, the sample containing protein biomarkers enters the channel and is exposed to laser light which is amplified by liquid crystal and a whispering-gallery-mode (WGM) laser emission spectrum is formed. The presence of protein causes a shift in the WGM spectrum, which is detected by a spectrometer.74
Exosome isolation and analysis
Exosomes are 30-150 nm sized extracellular vesicles secreted by cells into bodily fluids. These intercellular communication tools contain a variety of proteins, RNAs, DNA and lipids from the original cell. Therefore, exosomes can be considered as enriched information packages, the study of which provides valuable clinical information, assisting personalized diagnosis and therapy. The performance of conventional separation techniques is not adequate in separating exosomes from other vesicles and cells in the bodily fluids. Hence, microfluidic separation systems with higher resolution than other separation techniques are proposed. So far, various types of microfluidic systems have been developed for exosome isolation as reviewed in Table S1 in supplementary data.75 Isolation methods based on physical properties can also well be used to separate exosomes. For example, acoustic-based microfluidic systems can isolate exosomes in a blood sample based on size and shape. Porous membrane based microfluidic platforms also can simply separate microvesicles from biofluids. The diameter of these pores is optimized to only micro vesicles are allowed to pass, and the cells are trapped behind the filter. Performing an on-chip electrophoresis in the next step helps to better discretion of the microvesicles from the plasma proteins.75-77 Isolation methods based on immuno-affinity capture are one of the most routine separation methods. In these methods, a ligand is coated into the system's micro-channels to trap exosomes with a certain extracellular marker. This method is also used in cancer diagnosis due to this fact that exosomes carry significant amounts of cancer biomarkers. Research has shown that in addition to apoptotic cells, malignant cells and cancer derived microvesicles also contain large amounts of externalized phosphatidylserine (PS). Kang, et al. designed an ExoChip to isolate these exosomes containing externalized PS. This device has a large number of circular chambers containing coated annexin V protein which has a Ca2 + dependent affinity for PS. The large number of these trapping chambers implanted on this device increases its separation efficiency.78
Magnetic based microfluidic systems are also highly efficient for on-chip exosome isolation and analysis. In this approach, a specific ligand is attached to the magnetic beads and beads are mixed with the serum sample. The serum sample containing the beads enters the microchannels of the microfluidic device and by applying magnetic force and several washing steps, only the exosomes with the target biomarker remain. On-chip exosome analysis is performed by steps of fluorescence detection and TEM imaging.79,80
Drug screening
A recent discovery toward the human genome has discoveries toward the human genome has revolutionized diagnostic and therapeutic methods. The introduction of individual medicine has led to an attempt to carefully optimize therapies tailored to everyone’s genetic mechanism. This has also affected the pharmaceutical industry. Genetic diversity amongst individuals causes variant response to medications. Thus, drug screening is crucial especially for life threatening diseases like cancer.20,81 High-throughput, automation, inexpensiveness, and efficiency with low-sample volumes are important in drug screening. Drug screening microfluidic platforms, in addition to addressing these issues, allow for three-dimensional cell culture and drug screening in in-vivo-like conditions (Table 2).82,83
Table 2.
Microfluidic-based studies for drug screening
|
Drug
|
Microfluidic platform
|
References
|
| Doxorubicin and Cisplatin |
A device providing the possibility of studying different doses of the doxorubicin and cisplatin on tumor cell lines. Different drug-medium concentrations enter each chamber. |
84
|
| carboplatin, gemcitabine, capecitabine, topotecan, and navelbine |
A tow-part chip containing a CTC isolation and culture chamber, and six separate drug screen cavities. Due to the structure of the separation chamber, CTCs are separated from other blood cells based on their size and are trapped and expanded under non-adherent culture conditions. Then, by applying fluid flow, CTCs enter the drug screening chambers, and their viability is measured by luminescence analysis. |
85
|
| Doxorubicin |
A droplet system to create homotypic and heterotypic spheroids and investigate the effect of chemotherapeutic agents on these spheroids. The purpose of creating spheroids is to mimic the microenvironment of a tumor and to investigate the effects of cell interactions with each other and with the surrounding environment in response to treatment. |
86
|
Gemcitabine
Oxaliplatin
TNFα |
A braille valve chip capable of a single/combinatorial drug screening on very small cancer biopsy samples, in 48 hours. Two chemotherapy drugs for pancreatic cancer (Gemcitabine and Oxaliplatin), specific kinase inhibitor drugs, and one cytokine (TNFα) that activates apoptosis have been tested alone and in combinatorial in this device. |
87
|
| Anticoagulant drugs |
A bleeding chip with two orthogonal channels, one acts as a blood vessel, and the second is the bleeding channel. The hemostatic plug will form at the site of pillars. |
88
|
| 100 compounds and amphotericin B mixtures |
A chip with 20 units which each unit has micro-channels with a diameter of 3 to 6 microns, so Candida albicans are trapped individually in these channels. Fluorescent microscopy was used to determine live and dead cells. |
89
|
Doxorubicin
and Tirapazamine |
A chip providing an oxygen tension gradient environment for cancer study. The MDA-MB-231cells are seeded in a fibrin gel (3D microenvironment) and gas channels apply the oxygen tension. The response of cancer cells to anti-cancer drugs in the hypoxic tumor microenvironment can be examined (Fig. 3). |
90
|
| Cisplatin |
A multi-channel device for selective trapping cuboidal-shaped micro dissected tissues and exposure to different drug doses |
91
|
This is important in two ways; First, the results of drug screening in laboratory animals cannot always be generalized to humans92 and second, 3D microenvironment simulation is effective in drug screening, especially in cancer. In the tumor microenvironment, various factors can impress the potency of the chemotherapy agents and for this reason microfluidic systems are advantageous options for drug screening.93-95 Determining the effective dose of the drug is vital for positive therapeutic response and a prescription inappropriate dose of the drug both increases the probability of side effects of the drug and may lead to drug resistance. Microfluidic devices can be used to apply drug concentration gradients to determine the effective dose, cell viability, and the period of drug influx. Different doses of the chemotherapy drugs with the cell culture medium enter the central inlets of the microfluidic system and the result of their efficacy is inspected.84,90 (Fig. 3).
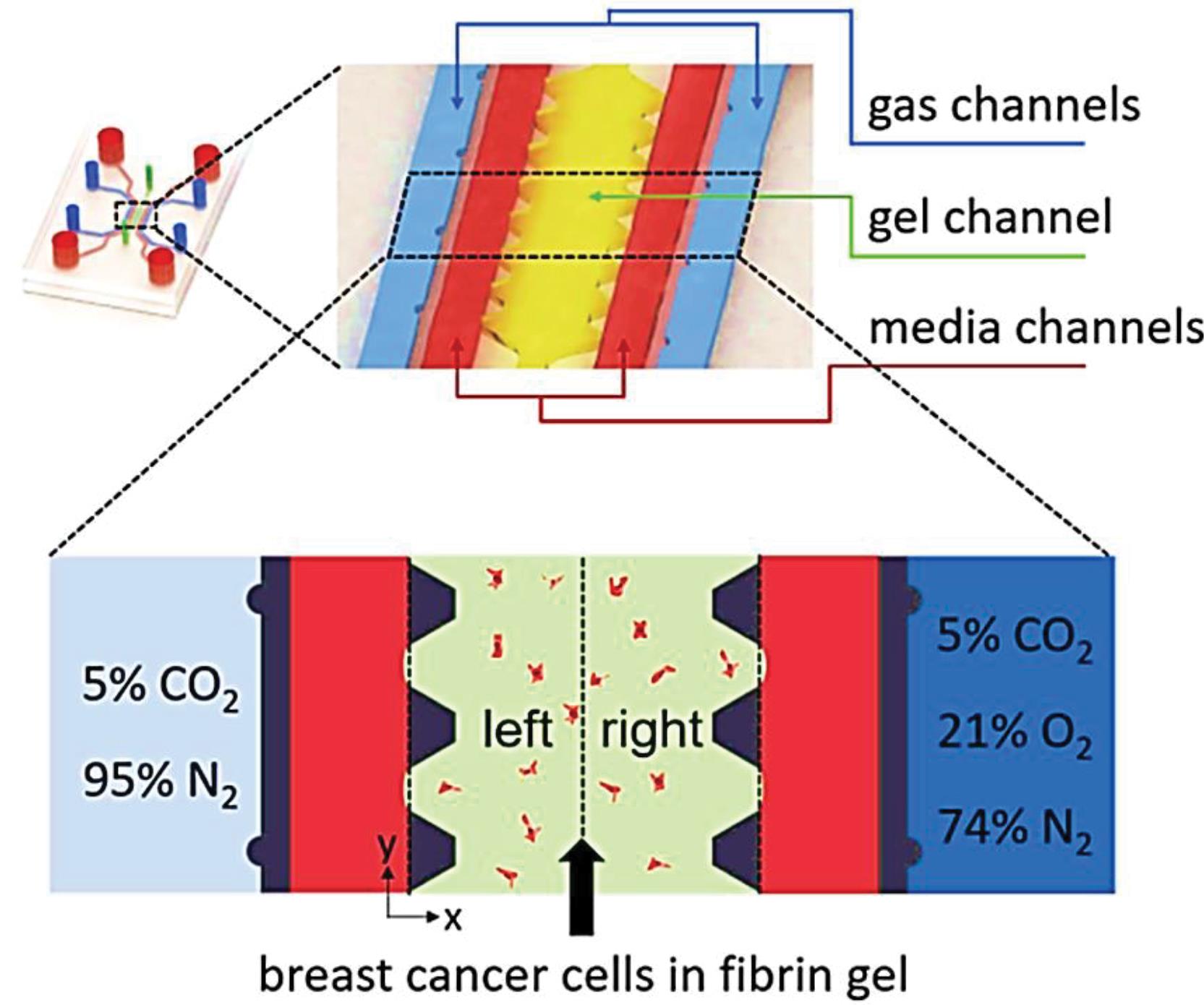
Fig. 3.
Schematic of an oxygen tension gradient providing device for studying the reaction of cancer cells to doxorubicin and tirapazamine in the hypoxic tumor microenvironment. Reprinted with permission from Namet al.90
.
Schematic of an oxygen tension gradient providing device for studying the reaction of cancer cells to doxorubicin and tirapazamine in the hypoxic tumor microenvironment. Reprinted with permission from Namet al.90
These multi-drug screening platforms make it possible to investigate the synergistic effect of several antibiotics and achieve an effective inhibitory dose.96 To prevent drug resistance, three-dimensional culture of biopsied samples on microfluidics and afterward drug screening have been performed.97 Eduati et al developed a braille valve chip capable of single/combinatorial drug screening on very small volumes of biopsy samples, in a period of 48 hours. Performing plug technology separation of samples using different chemical phases provided different test conditions on one chip at the same time. Moreover, several computer-controlled, movable pins contrived to precisely control drug circulation inside microchannel networks.87
Several microfluidic platforms (bleeding chips) have been designed to simulate in-vivo thrombosis condition98-101 for screening antithrombotic agents and determining the appropriate dose, even with a personalized treatment approach. Thrombosis is a pathophysiologic condition associated with a high rate of mortality worldwide correlated with many clinical complications including immune-mediated disease, cardiovascular disease, stroke, atherosclerosis, and malignancies.102 However, the urgent need for anticoagulant screening became even greater when the link between thrombosis and mortality from the global COVID-19 issue was identified.103 As the world sought to reduce the mortality rate of this pandemic, high throughput systems for drug screening came to the fore.88 The polydimethylsiloxane-based bleed chip designed by Lakshmanan, et al. is an example of these studies that two orthogonal channels embedded on this device. The main channel as the blood vessel and side-channel called the bleeding channel and three circular pillars located at the junction of two channels. The influx of whole blood into the channels of the bleeding chip with a determined flow rate led to the aggregation of platelet at the site of the pillars that led to the formation of the hemostatic plug caused blockage of the bleeding channel.88
Cancer study
Cancer is a heterogeneous disease described by uncontrolled cell proliferation and invasion104 that is the main health problem and a leading cause of death (millions of deaths each year) worldwide.105 However, the cancer mortality rate has decreased since the mid-1970s,105 but its prevalence is increasing due to changes in lifestyle such as smoking, lack of physical activity, and improper diets.106 Since early diagnosis of cancer is very important, different methods have been developed for detection and characterization of this disease. Nowadays, strategies for tumor diagnosis and treatment are critically influenced by the viewpoint of personalized medicine. A set of alterations at transcriptome, proteome, and metabolome levels are involved in cancer incidence, therefore, a molecular panel as a recognition pattern is necessary for prediction, diagnosis, and treatment of cancer. Predictive, preventive, and personalized medicine (PPPM) is an approach based on discovering key tumor molecule-panel for individuals and providing a treatment based on each person's own panel.107 In this regard, several microfluidic platforms have been developed for studying cancer biology, capturing cancer cells and subsequently enabling on-chip post-processing after capture, detecting cancer biomarkers, and testing drug sensitivity108-111 (Table 3; Fig. 4).
Table 3.
Cancer on a chip studies
|
Type of cancer
|
Microfluidic platform
|
Ref
|
| ovarian adenocarcinoma |
A middle micro channel surrounded by two lateral channel which is separated by array of PDMS column to loading of hydrogel for culturing of tumor spheroids and macrophage in separated channel. To create of tumor-microenvironment model, the tumor spheroid placed in collagen gel and macrophage were embedded in adjacent channel (Fig. 4A). |
110
|
| Leukemia-on-a-chip |
A device simulating leukemic bone-marrow niche and B-ALL. Three region including central venous sinus (central red part), a medullary cavity (green parts), and endosteal regions (external ring) are emulated in this device. These areas are divided by trapezoidal micro-columns containing cell-hydrogel mixture, balancing surface tension and capillary forces, to imitate the leukemic BM architecture. |
112
|
| Ovarian cancer on a chip |
A two-layared device with a separating thin, porous membrane imitating the microarchitecture of the tumor-vascular connection. Three ECM, tumor (in outer layer) and vascular (bottom layer) microchannels are separated by hexagonal micropillars which facilitates cell invasion. This device enables the study of intravenous perfusion of platelets, their penetration through the endothelium and cancer cells invasion. |
113
|
| Lung cancer-on-a-chip |
This chip includes a central chamber where the Hepatic stellate cells (HSCs) and hepatocellular carcinoma
(HCC) cells enter this chamber together with matrigel. Endothelial cells are cultured in the two side chambers and they play the role of blood vessels. Cellular crosstalk, Activation of HSCs, endothelial invasion, drug resistance and inefficiency of natural killer (NK) cells is studied on this chip. |
114
|
| Pancreatic cancer-on-a-chip |
A two-part microfluidic device was used to model the heterogeneous tumor microenvironment of pancreatic ductal adenocarcinoma. Patient-derived primary cancer cells (PCCs) along with stromal cells and macrophages were cultured in the upper chamber on the matrigel matrix. The lower chamber which is separated from the upper chamber by a permeable layer, provides the culture medium required for cell growth. |
115
|
| Kidney to liver metastasis-on-a-chip |
A poly (methyl methacrylate) (PMMA)/ PDMS fabricated device with 7 cell culture microwells containing different ratios of co-cultured immortalized hepatocytes HepLL and kidney cancer Caki-1 cells into an ECM of 2:3DLM/GelMA hydrogel to mimic the metastasis of kidney cancer to the liver (Fig. 4B). |
111
|
| breast-cancer-on-chip |
A tow-layered device which the upper layer contained three independent capillary vessel channels comprising an endothelial monolayer, connected ECM channel and a multicellular tumor spheroids (MCTS) culture channel with 11 U-shaped microchambers. Second layer was embedded under the U-shaped microchambers creating the perfusion flow along with preventing cells evasion. This system has been used for the assessment of drug delivery systems and toxicity studies. |
116
|
| Lung cancer on a chip |
A glass-based microfluidic device to investigate drug toxicity. This system is designed to provide a sensor integrated organoid for lung cancer monitoring and evaluation of the toxicity of candidate drugs. For real time monitoring the survival of cultured cells on glass-based chip, a trans-epithelial electrical (TEER) impedance sensor is embedded in this device. An optical pH sensor also is installed to check the pH of the culture medium. Therefore, this system has the ability to investigate the cell toxicity and determine the appropriate therapeutic dose |
117
|
| Tumor vessel-on-a-chip |
A MF device preparing the opportunity to imitate the formation of thrombi and hemorrhage in vascular tumors in vitro. Device is made of micro-sized hollow channels coated with fibronectin and endothelial cells. A microfluidic syringe pump has been installed to perfuse the blood at physiological flow rates and shear stress. |
118
|
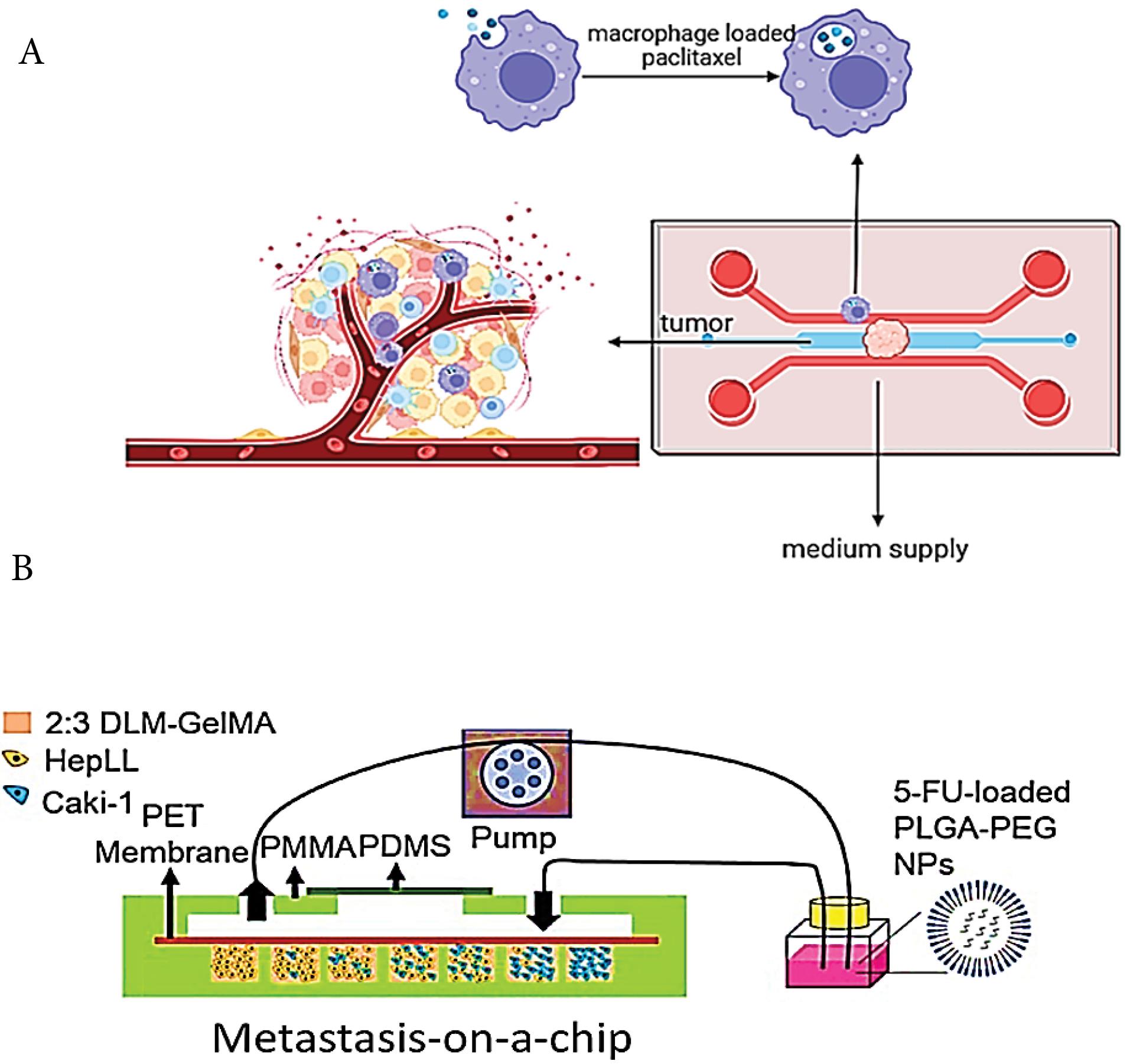
Fig. 4.
Schematics of microfluidic systems replicating cancers: (A) Ovarian adenocarcinoma,110 (B) Kidney to liver metastasis (Reprinted from Wang et al111 under the terms of the Creative Commons Attribution License; https://creativecommons.org/licenses/by/4.0/).
.
Schematics of microfluidic systems replicating cancers: (A) Ovarian adenocarcinoma,110 (B) Kidney to liver metastasis (Reprinted from Wang et al111 under the terms of the Creative Commons Attribution License; https://creativecommons.org/licenses/by/4.0/).
Although biopsy of tumor tissue is a common way of cancer diagnosis, due to its invasive nature, it is an inconvenient method for patients and can cause problems such as tissue damage. Studies have shown that individuals' body fluids contain biomarkers (DNA, RNA, protein and circulating tumor cells (that can be used in cancer detection and treatment follow up (based on each person's specific biomarkers).119 Microfluidic is a non-invasive, quick and suitable method for in vitro studying of individual’s body fluids.120 Paper-based systems can be mentioned among the tools provided. Paper-based microfluidics is one of the diagnostic tools that have found wide application in the field of biosensors due to their cheapness and ease of use. Wang et al developed a four-layer paper microfluidic system for the simultaneous detection of carcinoembryonic antigen (CEA) and neuron-specific enolase (NSE) cancer biomarkers. The diagnostic electrochemical system of this instrument has three electrodes and two Au-NP-based nanocomposites applied to immobilize the aptamers and intensify the electron transfer. The advantages of this system include sensitivity, selective power, and high detection speed.121
At a tumor site, some of the tumor cells (circulating tumor cells; CTCs) shed into the bloodstream and are responsible for the spreading of cancer to distant sites that provide personalized information about the stage of the disease and the efficacy of treatment.122,123 Thus, CTCs isolation from patient blood offers a valuable biomarker for diagnosis, characterization, and monitoring. Because of the low circulating levels and short lifespan, CTC separation requires a rapid and sensitive method. In this regard, immune-affinity based techniques have been investigated for CTCs isolation but these methods with multiple steps are time-consuming and have a high risk of loss of target cells in the separation stages. Microfluidic technology is a promising approach for high throughput, and sensitive separation of CTCs comparing to other methods.124 One example of a microfluidic system for CTC separation is the three-part system designed by Wu, et al. In this chip, the sample first enters the detection section, and by applying a continuous flow (Deterministic lateral displacement structure), CTCs are separated from the rest of the peripheral blood cells and enter the second section for pure isolation by encounter with specific antibody.125
It is worth mentioning that one of the most important microfluidic applications in the field of oncology is providing a proper 3D microenvironment for the study of cancer biology, metastasis, and drug resistance. The interaction between malignant cells and tumor associated cells at the site of the tumor forms the tumor microenvironment (TME) which plays a crucial role in metastasis, angiogenesis, and chemo resistance. Microfluidic platforms offer highly controllable conditions through controlling the shear stress, microperfusion and cell-cell or cell-matrix communication for better mimicking the TME. Thus microfluidic propose suitable models for studying the physiological processes of cancer as well as a high throughput system for drug screening.126,127 Moreover, microfluidic technology is a valuable tool for the analysis of cancer cell invasion and metastasis. Recently, several microfluidic devices have been offered for studies of cancer cells migration128 such as: the Kühlbach et al chip for study of the trans-migration and invasion of tumor cells from blood vessels129; growth factor gradient based systems130; chemotaxis-based prompted migration system131; perfusable vascular structures as a tumor angiogenesis and drug delivery model132; and mechanical confinement systems for studying spontaneous migration of cancer cells.133
Organ on chip
In recent years, studies have shown that cell niches have a tremendous effect on cellular demeanor, consequently the behavior of cells in two-dimensional cultures is far different from three-dimensional conditions. On the other hand, in-vivo animal studies also have not been able to overcome the challenge of in-vivo modeling of diseases and therapeutic responses due to species differences. Therefore, in recent years, many researchers have focused on in-vitro regeneration of human body organs (Fig. 5).134,135
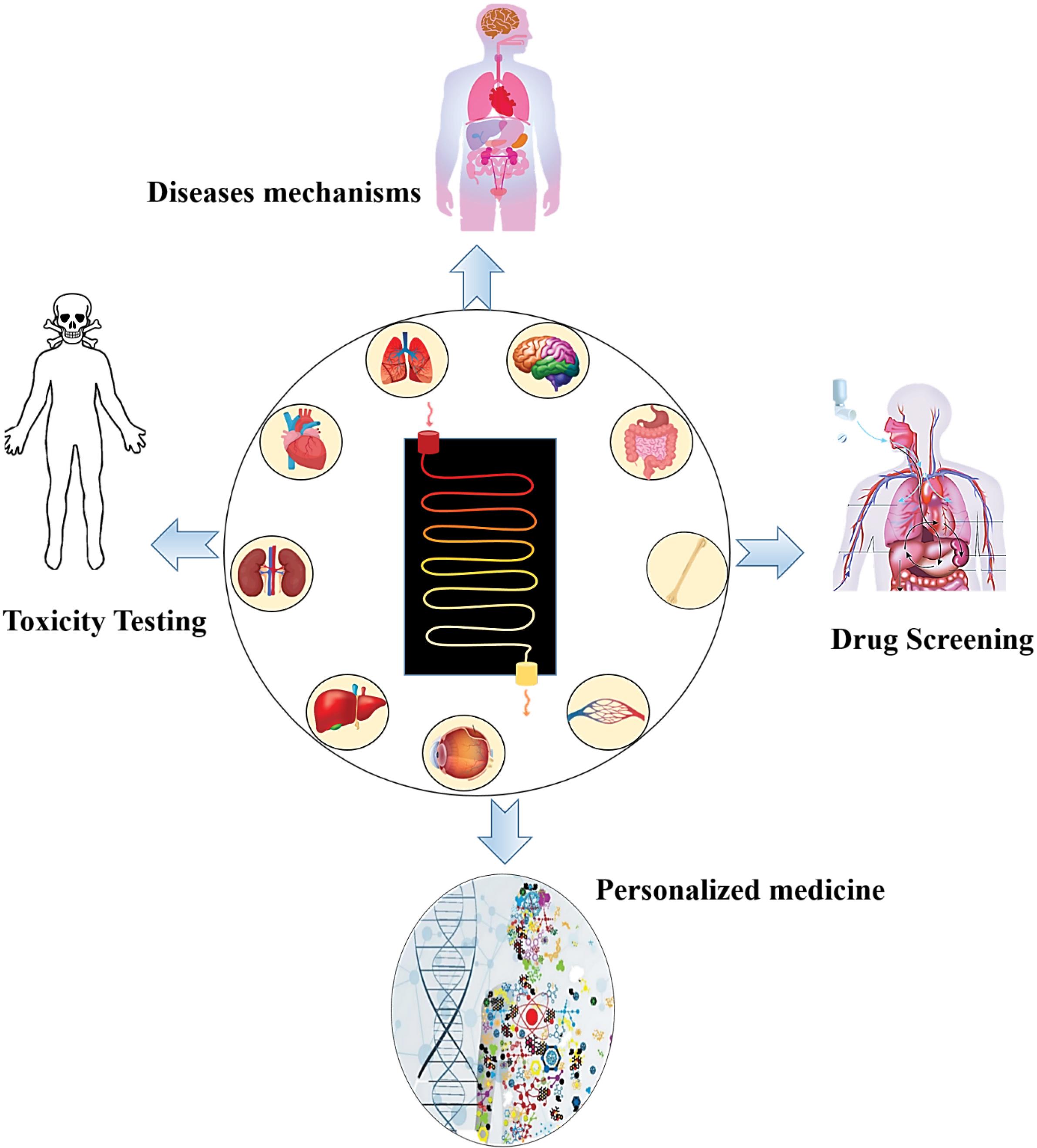
Fig. 5.
Applications of organ on a chip in biomedicine.
.
Applications of organ on a chip in biomedicine.
Organ-on-a-chip (OOC) is an approach in which cells in a microfluidic cultivation environment are cultured with controlled in-vivo like conditions. As a result, tissue or organ formed on these microfluidic devices are true simulations of human tissues/organs. OOC can be used for a variety medical purposes and personalization of treatment strategies where a specific therapy with more success probability and less side effects is required.136,137 So far, different types of human organs have been successfully remodeled on microfluidic platforms.135 For example, the lung on a chip is a reconstructed model of the lung that restores the function of this organ in an in-vitro condition. The physiology of this regenerated organ is similar to that of a normal organ, i.e. the epithelial and endothelial cells are arranged in a three-dimensional microenvironment similar to in-vivo conditions and perform the rhythmic function of respiration. These models can be used to study the pathological conditions of diseases, pharmacological and toxicology investigations with a personalized treatment perspective.138,139 Various platforms have been proposed to mimic lung function on the microfluidic system138,140,141 as shown in Table 4 and Fig. 6.
Table 4.
Organ-on-a -chip studies
|
Type of OOC
|
Cell source
|
Microfluidic platform
|
study
|
| Alveolar-capillary barrier-on-a-chip |
Alveolar epithelial cells,and endothelial cells |
This model consists of 3 parallel microchannels. The middle channel, which contains the matrigel imitating the extracellular matrix (ECM), separates the two side channels remodeling the alveolar and capillary structures. Alveolar epithelial cells are cultured on one side and endothelial cells on the other side. Nanoparticles exposed to epithelial cells and in the capillary side a fluidic flow similar to the human lung is applied. |
142
|
| Liver-on-a-chip |
Hepatocellular carcinoma HepG2, human hepatic stellate cell LX-2,and human umbilical vein endothelial cells (HUVECs) |
A system consists of endothelial cells and the permeable hydrogel to mimic diffusion. The microfluidic chip is consisted of two adjacent but discrete channels: inner channel which represent the capillary vessels, its surface is covered with the HUVECs and exposed with medium to show the shear stress in the physiological condition), outer channel: the outer channel is filled with the hepatic spheroid shapes which is formed by the concaved micro wells. The nutrient from inner channel could uptake through the infusion by the hydrogel. |
143
|
| Liver-kidney-on-a-chip |
HepG2,and Hek293 cells |
A microfluidic device consists of two connected chambers simulating kidney and liver function. Using an optimal flow velocity, the conditions for evaluating the primary and secondary toxic effects of drug candidates on liver and kidney are provided, respectively. |
144
|
| Kidney-liver -on-a-chip (an acute renal injury model) |
Kidney tubuloid and liver biopsies |
The aim of this study was to investigate the therapeutic effect of Mesenchymal stromal cell (MSC)-derived small extracellular vesicles (sEVs) on the repair of kidney damage, therefore, the modeling of kidney and liver organoids (accumulation place of sEVs) was done on a microfluidic system with two interconnected chambers. To create a functional tubular epithelium detaching the blood and urine, kidney compartment was cultured on a semipermeable membrane and kidney injury was caused by hydrogen peroxide. |
145
|
| Stomach-on-a-chip |
Human-derived adenocarcinoma epithelial cell line MKN74, and human stomach normal fibroblast cell line NST-20 |
This chip is a nine-part structure mimicking three different layers of the gastric mucosa; the epithelial barrier, the basement membrane made of fibronectin thin gel, and the lamina propria layer comprising collagen type I gel and normal gastric fibroblasts. A peristalsis-like motion is also applied to the system to reconstruct the dynamic stomach microenvironment. |
146
|
| Lung-on-a-chip |
Human alveolar epithelial cells (HPAEpiCs),
Human umbilical vein endothelial cells (HUVECs) |
A breathing alveolar–capillary chip consisting of the alveolar epithelial cells cultured in the upper side and pulmonary microvascular endothelial cells in the bottom side of the elastic membrane. Applying an air flow induce epithelial cells differentiation (Fig. 6A). |
140,147
|
| Vessel-on-a-chip |
human umbilical vein endothelial cells (HUVECs) |
A coaxial flow microfluidic device with a helical microfiber for fabricating different vessel structures such as multilayer, hollow. Coaxial flows are consisting of an inner alginate flow and outer CaCl2 flow (outer and inner flows are shifted in hollow microfibers). Two- and three-layer structures are also made by increasing the number of infusion micro-channels. HUVECs were seeded in the inner layer of the constructed membrane forming the endothelium layer as the lumen of blood vessels. |
148
|
| Heart-on-a-chip |
Human induced pluripotent stem (hiPS) cells derived cardiac cells |
Cardiac micro-tissues derived from human iPSCs cultured on a micro-electro-mechanical microfluidic system, comprising of a microchannel, a push bar placed on the top, a chamber and diaphragm. The cardiac cells are cultured on the push bar within a fibronectin matrix. Inside the microchannel, a culture medium containing fluorescent particles flows. Instinctive pulse generated by cardiac micro tissue can be measured by fluorescent particle displacement (Fig. 6B). |
141
|
| Heart-on-a-chip |
Ventricular cardio myocytes (CMs) were derived from the human embryonic stem cell (hESC),and the human induced pluripotent stem cell (hiPSC) |
A rectangular micro-well interconnected by two parallel grooves containing Poly (octamethylene maleate (anhydride) citrate) (POMaC) polymer wires. POMaC wires have an inherent fluorescence leading to monitoring the passive and active tension of cardiac tissue. |
149
|
| Brain-on-a-chip |
Human induced pluripotent stem (hiPS) cells |
A microfluidic system with two parallel chambers for cell culture and three separate channels. hiPS derived EBs mixed with matrigel are cultured in two parallel channels. The lateral channels contain the culture medium. The middle micropillar-shaped channel also contains the culture medium, causes fluid flow perfusion, and facilitates the exchange of oxygen and nutrients to cell-containing channels. |
150
|
| Kidney-on-a-chip |
Human induced pluripotent stem (hiPS) cells |
A device consists of a stretchable PDMS elastomer that comprise two parallel channels with opposite directions and segregated by a porous flexible PDMS membrane that its coated by laminin 511. |
151
|
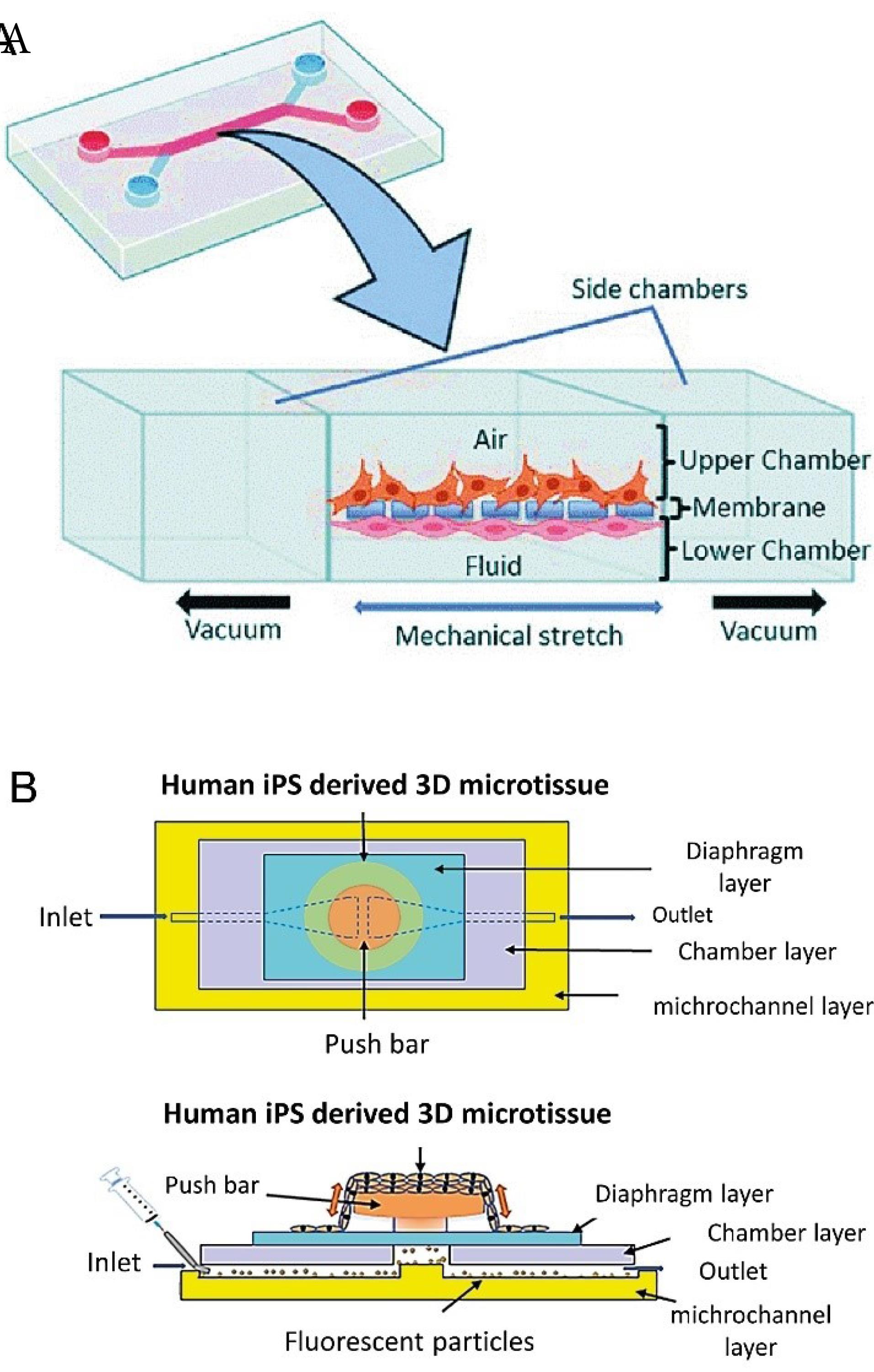
Fig. 6.
(A). The design of breathing alveolar–capillary chip, the alveolar epithelial cells are cultured in the upper side and pulmonary microvascular endothelial cells in the bottom side of the elastic membrane. Applying an air flow induce epithelial cells differentiation140 (B). Schematic of a cardiac micro-tissues derived from human iPSCs (Reprinted from Abulaiti et al141 under a Creative Commons Attribution 4.0 International License).
.
(A). The design of breathing alveolar–capillary chip, the alveolar epithelial cells are cultured in the upper side and pulmonary microvascular endothelial cells in the bottom side of the elastic membrane. Applying an air flow induce epithelial cells differentiation140 (B). Schematic of a cardiac micro-tissues derived from human iPSCs (Reprinted from Abulaiti et al141 under a Creative Commons Attribution 4.0 International License).
In lung organoid-based toxicological studies, a new agent/drug is exposed to remodeled organoid and the effect on tissue function is investigated. Nanoparticles (NPs) have recently been used in various industrial and clinical fields, therefore, the study of their effects on various human organs is vital. The formation of reactive oxygen species and the subsequent inflammatory responses are among the side effects of NPs. Some NPs also can cross body barriers such as the alveolar-capillary barrier and penetrate different parts of the body. Zhang et al developed a microfluidic device simulating the alveolar-capillary barrier function for assessment of NPs toxicity.40
Given the vital role of the liver in metabolism and detoxification, it is valuable to study of the physiology of liver disorders and discovery of effective drugs. In recent years, advances in liver organoids have greatly assisted to provide a more informative model to upgrade therapies toward more efficient, targeted, and personalized treatments.152 Liver disease accounts for 3.5% of global deaths and about 2 million deaths are recorded annually due to these diseases. Cirrhosis, hepatitis and hepatocellular carcinoma are among the most important liver diseases.153 Up to now, various two-dimensional (2D) and three-dimensional (3D) liver models have been reconstructed on the microfluidic system.154 An important challenge in liver regeneration is the reconstruction of the tubular structure and the provision of conditions for nutrient transfer between hepatocytes and vascular endothelial cells. To address this limitation, Meng et al fabricated a pluronic F127 (F127-DA) hydrogel microfluidic device, imitating the structural and functional properties of human liver. This F127-DA fabricated chip is able to afford high channel resolution along with tolerating the perfusion state. The hepatic spheroids are in the outer microchannel and the inner microchannel is covered with a single layer of endothelial cells. The culture medium at a flow rate similar to the physiological conditions is pumped amid these two channels. This in vitro 3D liver model expresses the potential for a range of applications including drug discovery and cellular toxicity analysis.143
In addition to studies on liver regeneration, studies have also examined the interaction of the liver with other organs. Remodeling of these interactions helps to study physiologic processes and related diseases. Reconstruction of the liver-gut model has a researching value to study metabolic routes and transmission of gut-absorbed lipids to the liver. This model can also be used to study the hepatic steatosis and drug screening for this prevalent disorder. To remodel the gut-liver interaction on the chip, a microfluidic device with two anti-parallel micro channels with opposite inputs and outputs was designed. Gut cells were cultured in the upper microchannel and hepatocytes were cultured in the lower microchannel. These channels are connected in the center, in which a permeable membrane was placed to exchange lipids. Screening of anti-steatotic agents on this system showed that compounds which help strengthen the gut barrier function, have a desirable outcome in treating hepatic steatosis.43
In addition to liver disease, heart diseases also have a high prevalence and the lethality risk. Thus, the reconstruction of an in vitro functional model of heart has been considered in recent years. These models have applications in physiological, pathological and drug screening studies. Whereas, the physical characteristics of the culture microenvironment, such as geometry and stiffness determine the differentiation and functionality of cardiac cells, the embedding of these conditions on a chip positively affect the outcome of the remodeling process.155 In addition to providing a suitable microenvironment for cell differentiation, the application of an adjusted electrical force is required, to induce cardiac cell contraction. Monitoring the active force and passive tension of cardiac cells also importantly authorizes personalized drug screening and toxicology studies.149,156 Moreover, the evaluation of the action potential presents vital information on the pathophysiologic state of cardiac cells. Liu, et al. developed a bioelectronic chip to study cardiac function under the hypoxia condition. The design of this micro-fluidic device is such that the input oxygen content is controlled by micro-channels so that embedded electrodes and platinum nanoparticles provide real-time monitoring of electrophysiological response of cardiac cells.157
The brain on a chip is perhaps one of the most important achievements of the microfluidic system. Although we have not been able to fully regenerate the brain due to the complex structure and technical limitations, successful regeneration of some parts and modeling of nervous system disorders has been promising for personalized treatment of neurological disorders. The human brain is a layer structure made up of different cells, including neurons, astrocytes, microglia, and oligodendrocytes. Neurons play a role in brain function through the establishment of synapses.158 Studies have shown that the neuron's microenvironment including adjacent cells and ECM affects its function. Neurons isolated from different parts of the brain demonstrate differences in metabolism, protein expression, and electrical activity. Therefore, an important challenge in in-vitro brain reconstruction is to provide this microenvironment to mimic the function of the target region in the brain.159 Interestingly, Park et al designed a microfluidic system for the simultaneous culture of neurons, astrocytes, and microglia cells and subsequently investigated the cellular interactions and mechanism of neuronal cell destruction during Alzheimer's disease. This 3D model is capable of recapitulating beta-amyloid aggregation, phosphorylated tau accumulation, neuroinflammatory activity, recruitment of microglia and cellular damage mediated by microglia cells.160 The study of cell communication is also important in the incidence of secondary neuronal cell death. Released glutamate from damaged cells, as well as synaptic connections, are involved in the spread of neuronal damage at the stroke or injury region. Samson, et al designed a microfluidic device with five parallel interconnected culture chambers for investigation the expansion of neurotoxicity. These chambers are designed to study the spread of cytotoxicity to adjacent regions through synapses. Therefore, in this model, synaptic dependent hyperactivity, sudden ion influx into neurons causing depolarization and cytotoxicity due to glutamate accumulation can be simulated. The results of this study revealed a protective mechanism to prevent the spread of cytotoxicity. In the era adjacent to the site of injury, signaling of the GluN2A receptor triggers the recruitment of GABA receptor, thereby hindering spreading cytotoxicity. These results could open new horizons for the treatment of progressive brain injuries.161 Blood-brain barrier (BBB) modeling is another important study in the area of the brain on a chip. BBB by controlling the entry of substances into nerve cells is a competent research option for the studies of diseases mechanism as well as drug delivery. Several CNS diseases and even neurological malignancies such as glioblastoma are associated with impaired BBB permeability. Due to the complexity of this system as well as species differences, 3D modeling of BBB on a chip can be superior to animal studies and 2D culture systems.162,163 In a recent study, Yu, et al. designed a microfluidic platform mimicking the BBB structural complexity and function. This model imitator the interactions between endothelial cells (ECs), pericytes, and surrounding astrocytes in BBB organization and the fluid flow condition in the brain microvasculature. One inlet, one outlet and a microchannel are embedded in this microfluidic device, and the inlet and outlet heights are designed in such a way that the fluid moves towards the outlet by the siphoning effect and via gravity. To slow down the flow speed as blood flow in brain microvasculature, a paper cylinder flow resistor installed in the outlet reservoir. The microchannel is also made in the form of a hollow lumen of type Ⅰ collagen hydrogel as a matrix for 3D cell culture. Yu, et al. then used TNF‐α to induce the inflammatory condition modifying the BBB permeability, as is the case with many CNS diseases. This device can provide a suitable drug screening platform for the treatment of neurovascular disorders.164
Concluding remarks
Given the valuable advances in biomedical science in recent decades, as well as the increasing knowledge of disease mechanisms, it is expected that personalized medicine will replace traditional methods of treatment in the not-too-distant future.20 This evolution certainly requires reformation in existing tools in the field of diagnosis, disease monitoring, and especially treatment providing the ability to tracking small amounts of biomolecules or mimicking the complex conditions within the patient body.165 Microfluidic devises, which attracted the attention of biomedical researchers due to their high accuracy and automation, were able to address many of the challenges in increasing the speed and accuracy of diagnostic methods as well as personalization of therapeutic approaches.166 A variety of microfluidic platforms are offered in biomedical applications and due to the advantages of these devices over traditional techniques, microfluidic technology is becoming a valuable portion in the field of personalized medicine.43 Point-of-care (POC) diagnostics,167 body on a chip, disease modeling, and rapid drug screening systems are all valuable microfluidic achievements in biomedicine.135 Increasing advances in the design and construction of microfluidic systems in these areas will be promising for further advances in the early diagnosis and the personalization of treatment methods.
Review Highlights
What is the current knowledge?
-
Genetic differences in individuals will affect their response to drugs and treatments.
-
Microfluidic instruments have the ability to accurately control the internal conditions of the system, with use in biological studies.
What is new here?
-
Different types of microfluidic systems with the ability to perform molecular tests have been invented for the purpose of quick and early diagnosis of diseases.
-
Microfluidic systems with 3D cell culture capability are designed to regenerate organs and diseases in laboratory conditions.
-
Organoid models allow for novel drug elucidation and researches in various human disorders.
Acknowledgments
The authors would like to thank the Deputy of the Research and Technology, ZUMS.
Competing Interests
None declared.
Ethical Statement
None to be declared.
References
- Bragheri F, Martínez Vázquez R, Osellame R. Microfluidics. In: Baldacchini T, ed. Three-Dimensional Microfabrication Using Two-Photon Polymerization. 2nd ed. William Andrew Publishing; 2020. p. 493-526. 10.1016/b978-0-12-817827-0.00057-6.
- Chen W, Shao F, Xianyu Y. Microfluidics-implemented biochemical assays: from the perspective of readout. Small 2020; 16:e1903388. doi: 10.1002/smll.201903388 [Crossref] [ Google Scholar]
- Zhang Q, Feng S, Lin L, Mao S, Lin JM. Emerging open microfluidics for cell manipulation. Chem Soc Rev 2021; 50:5333-48. doi: 10.1039/d0cs01516d [Crossref] [ Google Scholar]
- Shakeri A, Khan S, Didar TF. Conventional and emerging strategies for the fabrication and functionalization of PDMS-based microfluidic devices. Lab Chip 2021; 21:3053-75. doi: 10.1039/d1lc00288k [Crossref] [ Google Scholar]
- Akther F, Yakob SB, Nguyen NT, Ta HT. Surface modification techniques for endothelial cell seeding in PDMS microfluidic devices. Biosensors (Basel) 2020; 10:182. doi: 10.3390/bios10110182 [Crossref] [ Google Scholar]
- Kemkemer R, Zenghao Z, Linxiao Y, Athanasopulu K, Frey K, Cui Z. Surface modification of polydimethylsiloxane by hydrogels for microfluidic applications. Curr Dir Biomed Eng 2019; 5(1):93-6. doi: 10.1515/cdbme-2019-0024 [Crossref] [ Google Scholar]
- Jensen C, Teng Y. Is it time to start transitioning from 2D to 3D cell culture? Front Mol Biosci 2020. 7: 33. 10.3389/fmolb.2020.00033.
- Kapałczyńska M, Kolenda T, Przybyła W, Zajączkowska M, Teresiak A, Filas V. 2D and 3D cell cultures - a comparison of different types of cancer cell cultures. Arch Med Sci 2018; 14:910-9. doi: 10.5114/aoms.2016.63743 [Crossref] [ Google Scholar]
- Verjans ET, Doijen J, Luyten W, Landuyt B, Schoofs L. Three-dimensional cell culture models for anticancer drug screening: worth the effort?. J Cell Physiol 2018; 233:2993-3003. doi: 10.1002/jcp.26052 [Crossref] [ Google Scholar]
- Zhao Q, Cui H, Wang Y, Du X. Microfluidic platforms toward rational material fabrication for biomedical applications. Small 2020; 16:e1903798. doi: 10.1002/smll.201903798 [Crossref] [ Google Scholar]
- Oliveira NM, Vilabril S, Oliveira MB, Reis RL, Mano JF. Recent advances on open fluidic systems for biomedical applications: a review. Mater Sci Eng C Mater Biol Appl 2019; 97:851-63. doi: 10.1016/j.msec.2018.12.040 [Crossref] [ Google Scholar]
- Coluccio ML, Perozziello G, Malara N, Parrotta E, Zhang P, Gentile F. Microfluidic platforms for cell cultures and investigations. Microelectron Eng 2019; 208:14-28. doi: 10.1016/j.mee.2019.01.004 [Crossref] [ Google Scholar]
- Wang A, Madden LA, Paunov VN. Advanced biomedical applications based on emerging 3D cell culturing platforms. J Mater Chem B 2020; 8:10487-501. doi: 10.1039/d0tb01658f [Crossref] [ Google Scholar]
- Hassan M, Awan FM, Naz A, de Andrés-Galiana EJ, Alvarez O, Cernea A. Innovations in genomics and big data analytics for personalized medicine and health care: a review. Int J Mol Sci 2022; 23:4645. doi: 10.3390/ijms23094645 [Crossref] [ Google Scholar]
- Goetz LH, Schork NJ. Personalized medicine: motivation, challenges, and progress. Fertil Steril 2018; 109:952-63. doi: 10.1016/j.fertnstert.2018.05.006 [Crossref] [ Google Scholar]
- Pasche B, Absher D. Whole-genome sequencing: a step closer to personalized medicine. JAMA 2011; 305:1596-7. doi: 10.1001/jama.2011.484 [Crossref] [ Google Scholar]
- Oates JT, Lopez D. Pharmacogenetics: an important part of drug development with a focus on its application. Int J Biomed Investig 2018; 1:111. doi: 10.31531/2581-4745.1000111 [Crossref] [ Google Scholar]
- Green ED, Gunter C, Biesecker LG, Di Francesco V, Easter CL, Feingold EA. Strategic vision for improving human health at The Forefront of Genomics. Nature 2020; 586:683-92. doi: 10.1038/s41586-020-2817-4 [Crossref] [ Google Scholar]
- Liggett SB. Pharmacogenetic applications of the human genome project. Nat Med 2001; 7:281-3. doi: 10.1038/85411 [Crossref] [ Google Scholar]
- Ginsburg GS, McCarthy JJ. Personalized medicine: revolutionizing drug discovery and patient care. Trends Biotechnol 2001; 19:491-6. doi: 10.1016/s0167-7799(01)01814-5 [Crossref] [ Google Scholar]
- Lauschke VM, Ingelman-Sundberg M. How to consider rare genetic variants in personalized drug therapy. Clin Pharmacol Ther 2018; 103:745-8. doi: 10.1002/cpt.976 [Crossref] [ Google Scholar]
- Goričar K, Dolžan V, Lenassi M. Extracellular vesicles: a novel tool facilitating personalized medicine and pharmacogenomics in oncology. Front Pharmacol 2021; 12:671298. doi: 10.3389/fphar.2021.671298 [Crossref] [ Google Scholar]
- Hassan R, Allali I, Agamah FE, Elsheikh SSM, Thomford NE, Dandara C. Drug response in association with pharmacogenomics and pharmacomicrobiomics: towards a better personalized medicine. Brief Bioinform 2021; 22:bbaa292. doi: 10.1093/bib/bbaa292 [Crossref] [ Google Scholar]
- Iriart JAB. Precision medicine/personalized medicine: a critical analysis of movements in the transformation of biomedicine in the early 21st century. Cad Saude Publica 2019; 35:e00153118. doi: 10.1590/0102-311x00153118 [Crossref] [ Google Scholar]
- Liu F, Huang J, Ning B, Liu Z, Chen S, Zhao W. Drug discovery via human-derived stem cell organoids. Front Pharmacol 2016; 7:334. doi: 10.3389/fphar.2016.00334 [Crossref] [ Google Scholar]
- Ingber DE. Human organs-on-chips for disease modelling, drug development and personalized medicine. Nat Rev Genet 2022; 23:467-91. doi: 10.1038/s41576-022-00466-9 [Crossref] [ Google Scholar]
- Yu F, Hunziker W, Choudhury D. Engineering microfluidic organoid-on-a-chip platforms. Micromachines (Basel) 2019; 10:165. doi: 10.3390/mi10030165 [Crossref] [ Google Scholar]
- Unagolla JM, Jayasuriya AC. Recent advances in organoid engineering: a comprehensive review. Appl Mater Today 2022; 29:101582. doi: 10.1016/j.apmt.2022.101582 [Crossref] [ Google Scholar]
- Burklund A, Tadimety A, Nie Y, Hao N, Zhang JXJ. Advances in diagnostic microfluidics. Adv Clin Chem 2020; 95:1-72. doi: 10.1016/bs.acc.2019.08.001 [Crossref] [ Google Scholar]
- Easley CJ, Karlinsey JM, Bienvenue JM, Legendre LA, Roper MG, Feldman SH. A fully integrated microfluidic genetic analysis system with sample-in-answer-out capability. Proc Natl Acad Sci U S A 2006; 103:19272-7. doi: 10.1073/pnas.0604663103 [Crossref] [ Google Scholar]
- Yang Y, Chen Y, Tang H, Zong N, Jiang X. Microfluidics for biomedical analysis. Small Methods 2020; 4:1900451. doi: 10.1002/smtd.201900451 [Crossref] [ Google Scholar]
- Wu Y, Wang Y, Lu Y, Luo X, Huang Y, Xie T. Microfluidic technology for the isolation and analysis of exosomes. Micromachines (Basel) 2022; 13:1571. doi: 10.3390/mi13101571 [Crossref] [ Google Scholar]
- Zhong Q, Ding H, Gao B, He Z, Gu Z. Advances of microfluidics in biomedical engineering. Adv Mater Technol 2019; 4:1800663. doi: 10.1002/admt.201800663 [Crossref] [ Google Scholar]
- Dwivedi S, Purohit P, Misra R, Pareek P, Goel A, Khattri S. Diseases and molecular diagnostics: a step closer to precision medicine. Indian J Clin Biochem 2017; 32:374-98. doi: 10.1007/s12291-017-0688-8 [Crossref] [ Google Scholar]
- Jayamohan H, Sant HJ, Gale BK. Applications of microfluidics for molecular diagnostics. Methods Mol Biol 2013; 949:305-34. doi: 10.1007/978-1-62703-134-9_20 [Crossref] [ Google Scholar]
- Abdulbaqi IM, Abou Assi R, Yaghmur A, Darwis Y, Mohtar N, Parumasivam T. Pulmonary delivery of anticancer drugs via lipid-based nanocarriers for the treatment of lung cancer: an update. Pharmaceuticals (Basel) 2021; 14:725. doi: 10.3390/ph14080725 [Crossref] [ Google Scholar]
- Purohit B, Vernekar PR, Shetti NP, Chandra P. Biosensor nanoengineering: design, operation, and implementation for biomolecular analysis. Sens Int 2020; 1:100040. doi: 10.1016/j.sintl.2020.100040 [Crossref] [ Google Scholar]
- Liu X, Wu W, Cui D, Chen X, Li W. Functional micro-/nanomaterials for multiplexed biodetection. Adv Mater 2021; 33:e2004734. doi: 10.1002/adma.202004734 [Crossref] [ Google Scholar]
- Soares RR, Neumann F, Caneira CR, Madaboosi N, Ciftci S, Hernández-Neuta I. Silica bead-based microfluidic device with integrated photodiodes for the rapid capture and detection of rolling circle amplification products in the femtomolar range. Biosens Bioelectron 2019; 128:68-75. doi: 10.1016/j.bios.2018.12.004 [Crossref] [ Google Scholar]
- Yin C, Wang Y, Ji J, Cai B, Chen H, Yang Z. Molecular profiling of pooled circulating tumor cells from prostate cancer patients using a dual-antibody-functionalized microfluidic device. Anal Chem 2018; 90:3744-51. doi: 10.1021/acs.analchem.7b03536 [Crossref] [ Google Scholar]
- Chiavaioli F, Santano Rivero D, Del Villar I, Socorro‐Leránoz AB, Zhang X, Li K. Ultrahigh sensitive detection of Tau protein as Alzheimer's biomarker via microfluidics and nanofunctionalized optical fiber sensors. Adv Photonics Res 2022; 3:2200044. doi: 10.1002/adpr.202200044 [Crossref] [ Google Scholar]
- Chinnamani MV, Hanif A, Kannan PK, Kaushal S, Sultan MJ, Lee NE. Soft microfiber-based hollow microneedle array for stretchable microfluidic biosensing patch with negative pressure-driven sampling. Biosens Bioelectron 2023; 237:115468. doi: 10.1016/j.bios.2023.115468 [Crossref] [ Google Scholar]
- Lee SY, Sung JH. Gut-liver on a chip toward an in vitro model of hepatic steatosis. Biotechnol Bioeng 2018; 115:2817-27. doi: 10.1002/bit.26793 [Crossref] [ Google Scholar]
- Sardesai NP, Kadimisetty K, Faria R, Rusling JF. A microfluidic electrochemiluminescent device for detecting cancer biomarker proteins. Anal Bioanal Chem 2013; 405:3831-8. doi: 10.1007/s00216-012-6656-5 [Crossref] [ Google Scholar]
- Yadav SS, Sikarwar BS, Ranjan P, Janardhanan R. Microfluidic system for screening disease based on physical properties of blood. Bioimpacts 2020; 10:141-50. doi: 10.34172/bi.2020.18 [Crossref] [ Google Scholar]
- Arata H, Komatsu H, Hosokawa K, Maeda M. Rapid and sensitive microRNA detection with laminar flow-assisted dendritic amplification on power-free microfluidic chip. PLoS One 2012; 7:e48329. doi: 10.1371/journal.pone.0048329 [Crossref] [ Google Scholar]
- Chen JS, Chen PF, Lin HT, Huang NT. A Localized surface plasmon resonance (LSPR) sensor integrated automated microfluidic system for multiplex inflammatory biomarker detection. Analyst 2020; 145:7654-61. doi: 10.1039/d0an01201g [Crossref] [ Google Scholar]
- Kulkarni MB, Goel S. Advances in continuous-flow based microfluidic PCR devices—a review. Eng Res Express 2020; 2:042001. doi: 10.1088/2631-8695/abd287 [Crossref] [ Google Scholar]
- Li Z, Bai Y, You M, Hu J, Yao C, Cao L. Fully integrated microfluidic devices for qualitative, quantitative and digital nucleic acids testing at point of care. Biosens Bioelectron 2021; 177:112952. doi: 10.1016/j.bios.2020.112952 [Crossref] [ Google Scholar]
- Shin S, Han D, Park MC, Mun JY, Choi J, Chun H. Separation of extracellular nanovesicles and apoptotic bodies from cancer cell culture broth using tunable microfluidic systems. Sci Rep 2017; 7:9907. doi: 10.1038/s41598-017-08826-w [Crossref] [ Google Scholar]
- Park J, Han DH, Hwang SH, Park JK. Reciprocating flow-assisted nucleic acid purification using a finger-actuated microfluidic device. Lab Chip 2020; 20:3346-53. doi: 10.1039/d0lc00432d [Crossref] [ Google Scholar]
- Kim CH, Park J, Kim SJ, Ko DH, Lee SH, Lee SJ. On-site extraction and purification of bacterial nucleic acids from blood samples using an unpowered microfluidic device. Sens Actuators B Chem 2020; 320:128346. doi: 10.1016/j.snb.2020.128346 [Crossref] [ Google Scholar]
- Ramos-Payán M, Ocaña-Gonzalez JA, Fernández-Torres RM, Llobera A, Bello-López M. Recent trends in capillary electrophoresis for complex samples analysis: A review. Electrophoresis 2018; 39:111-25. doi: 10.1002/elps.201700269 [Crossref] [ Google Scholar]
- Sun Y. Microfluidic capillary electrophoresis chip techniques: theory and different separation modes. In: Li XJ, Yang C, Li PC, eds. Multidisciplinary Microfluidic and Nanofluidic Lab-on-a-Chip. Amsterdam: Elsevier; 2022. p. 99-142. 10.1016/b978-0-444-59432-7.00002-9.
- Farmerie L, Rustandi RR, Loughney JW, Dawod M. Recent advances in isoelectric focusing of proteins and peptides. J Chromatogr A 2021; 1651:462274. doi: 10.1016/j.chroma.2021.462274 [Crossref] [ Google Scholar]
- Araya-Farias M, Dziomba S, Tran NT. Microfluidic strategies for extraction and preconcentration of proteins and peptides. In: Hussain CM, ed. Handbook on Miniaturization in Analytical Chemistry. Elsevier; 2020. p. 35-75. 10.1016/b978-0-12-819763-9.00003-9.
- Lee Y, Kwon JS. Microfluidic free-flow electrophoresis: a promising tool for protein purification and analysis in proteomics. J Ind Eng Chem 2022; 109:79-99. doi: 10.1016/j.jiec.2022.02.028 [Crossref] [ Google Scholar]
- Molinski J, Tadimety A, Burklund A, Zhang JXJ. Scalable signature-based molecular diagnostics through on-chip biomarker profiling coupled with machine learning. Ann Biomed Eng 2020; 48:2377-99. doi: 10.1007/s10439-020-02593-y [Crossref] [ Google Scholar]
- Shaw KJ, Birch C, Hughes EM, Jakes AD, Greenman J, Haswell SJ. Microsystems for personalized biomolecular diagnostics. Eng Life Sci 2011; 11:121-32. doi: 10.1002/elsc.201000175 [Crossref] [ Google Scholar]
- Pan LH, Pang ST, Fang PY, Chuang CK, Yang HW. Label-free biochips for accurate detection of prostate cancer in the clinic: dual biomarkers and circulating tumor cells. Theranostics 2017; 7:4289-300. doi: 10.7150/thno.21092 [Crossref] [ Google Scholar]
- Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science 2005; 309:1519-24. doi: 10.1126/science.1111444 [Crossref] [ Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A. RAS is regulated by the let-7 microRNA family. Cell 2005; 120:635-47. doi: 10.1016/j.cell.2005.01.014 [Crossref] [ Google Scholar]
- Dwivedi S, Purohit P, Sharma P. microRNAs and diseases: promising biomarkers for diagnosis and therapeutics. Indian J Clin Biochem 2019; 34:243-5. doi: 10.1007/s12291-019-00844-x [Crossref] [ Google Scholar]
- Condrat CE, Thompson DC, Barbu MG, Bugnar OL, Boboc A, Cretoiu D. miRNAs as biomarkers in disease: latest findings regarding their role in diagnosis and prognosis. Cells 2020; 9:276. doi: 10.3390/cells9020276 [Crossref] [ Google Scholar]
- Khashayar P, Al-Madhagi S, Azimzadeh M, Scognamiglio V, Arduini F. New frontiers in microfluidics devices for miRNA analysis. TrAC Trends Anal Chem 2022; 156:116706. doi: 10.1016/j.trac.2022.116706 [Crossref] [ Google Scholar]
- Lee H, Lee J, Lee SG, Doyle PS. Hydrogel-based colorimetric assay for multiplexed microRNA detection in a microfluidic device. Anal Chem 2020; 92:5750-5. doi: 10.1021/acs.analchem.9b05043 [Crossref] [ Google Scholar]
- Guo S, Lin WN, Hu Y, Sun G, Phan DT, Chen CH. Ultrahigh-throughput droplet microfluidic device for single-cell miRNA detection with isothermal amplification. Lab Chip 2018; 18:1914-20. doi: 10.1039/c8lc00390d [Crossref] [ Google Scholar]
- Sun X, Wang H, Jian Y, Lan F, Zhang L, Liu H. Ultrasensitive microfluidic paper-based electrochemical/visual biosensor based on spherical-like cerium dioxide catalyst for miR-21 detection. Biosens Bioelectron 2018; 105:218-25. doi: 10.1016/j.bios.2018.01.025 [Crossref] [ Google Scholar]
- Bellassai N, D'Agata R, Spoto G. Isothermal circular strand displacement-based assay for microRNA detection in liquid biopsy. Anal Bioanal Chem 2022; 414:6431-40. doi: 10.1007/s00216-022-04228-8 [Crossref] [ Google Scholar]
- Chircov C, Bîrcă AC, Grumezescu AM, Andronescu E. Biosensors-on-chip: an up-to-date review. Molecules 2020; 25:6013. doi: 10.3390/molecules25246013 [Crossref] [ Google Scholar]
- Rahman M, Islam KR, Islam MR, Islam MJ, Kaysir MR, Akter M. A critical review on the sensing, control, and manipulation of single molecules on optofluidic devices. Micromachines (Basel) 2022; 13:948. doi: 10.3390/mi13060968 [Crossref] [ Google Scholar]
- Psaltis D, Quake SR, Yang C. Developing optofluidic technology through the fusion of microfluidics and optics. Nature 2006; 442:381-6. doi: 10.1038/nature05060 [Crossref] [ Google Scholar]
- Rivet C, Lee H, Hirsch A, Hamilton S, Lu H. Microfluidics for medical diagnostics and biosensors. Chem Eng Sci 2011; 66:1490-507. doi: 10.1016/j.ces.2010.08.015 [Crossref] [ Google Scholar]
- Wang Z, Liu Y, Gong C, Yuan Z, Shen L, Chang P. Liquid crystal-amplified optofluidic biosensor for ultra-highly sensitive and stable protein assay. Photonix 2021; 2:18. doi: 10.1186/s43074-021-00041-1 [Crossref] [ Google Scholar]
- Contreras-Naranjo JC, Wu HJ, Ugaz VM. Microfluidics for exosome isolation and analysis: enabling liquid biopsy for personalized medicine. Lab Chip 2017; 17:3558-77. doi: 10.1039/c7lc00592j [Crossref] [ Google Scholar]
- Iliescu FS, Vrtačnik D, Neuzil P, Iliescu C. Microfluidic technology for clinical applications of exosomes. Micromachines (Basel) 2019; 10:392. doi: 10.3390/mi10060392 [Crossref] [ Google Scholar]
- Liga A, Vliegenthart AD, Oosthuyzen W, Dear JW, Kersaudy-Kerhoas M. Exosome isolation: a microfluidic road-map. Lab Chip 2015; 15:2388-94. doi: 10.1039/c5lc00240k [Crossref] [ Google Scholar]
- Kang YT, Purcell E, Palacios-Rolston C, Lo TW, Ramnath N, Jolly S. Isolation and profiling of circulating tumor-associated exosomes using extracellular vesicular lipid-protein binding affinity based microfluidic device. Small 2019; 15:e1903600. doi: 10.1002/smll.201903600 [Crossref] [ Google Scholar]
- Le MN, Fan ZH. Exosome isolation using nanostructures and microfluidic devices. Biomed Mater 2021; 16:022005. doi: 10.1088/1748-605X/abde70 [Crossref] [ Google Scholar]
- Lu Y, Cheng Z, Wang K, Qu Y, Bai Y, Qiu S, et al. Integrated on-Chip Isolation and Analysis of Exosome Tumor Markers via Microfluidic System. In: 2019 20th International Conference on Solid-State Sensors, Actuators and Microsystems & Eurosensors XXXIII (TRANSDUCERS & EUROSENSORS XXXIII). Berlin: IEEE; 2019. p. 956-9. 10.1109/transducers.2019.8808641.
- Li YY, Jones SJ. Drug repositioning for personalized medicine. Genome Med 2012; 4:27. doi: 10.1186/gm326 [Crossref] [ Google Scholar]
- Shi Y, Cai Y, Cao Y, Hong Z, Chai Y. Recent advances in microfluidic technology and applications for anti-cancer drug screening. TrAC Trends Anal Chem 2021; 134:116118. doi: 10.1016/j.trac.2020.116118 [Crossref] [ Google Scholar]
- Mitxelena-Iribarren O, Zabalo J, Arana S, Mujika M. Improved microfluidic platform for simultaneous multiple drug screening towards personalized treatment. Biosens Bioelectron 2019; 123:237-43. doi: 10.1016/j.bios.2018.09.001 [Crossref] [ Google Scholar]
- Shen S, Zhang X, Zhang F, Wang D, Long D, Niu Y. Three-gradient constructions in a flow-rate insensitive microfluidic system for drug screening towards personalized treatment. Talanta 2020; 208:120477. doi: 10.1016/j.talanta.2019.120477 [Crossref] [ Google Scholar]
- Schwab FD, Scheidmann MC, Ozimski LL, Kling A, Armbrecht L, Ryser T. MyCTC chip: microfluidic-based drug screen with patient-derived tumour cells from liquid biopsies. Microsyst Nanoeng 2022; 8:130. doi: 10.1038/s41378-022-00467-y [Crossref] [ Google Scholar]
- Lee SI, Choi YY, Kang SG, Kim TH, Choi JW, Kim YJ. 3D multicellular tumor spheroids in a microfluidic droplet system for investigation of drug resistance. Polymers (Basel) 2022; 14:3752. doi: 10.3390/polym14183752 [Crossref] [ Google Scholar]
- Eduati F, Utharala R, Madhavan D, Neumann UP, Longerich T, Cramer T. A microfluidics platform for combinatorial drug screening on cancer biopsies. Nat Commun 2018; 9:2434. doi: 10.1038/s41467-018-04919-w [Crossref] [ Google Scholar]
- Lakshmanan HHS, Pore AA, Kohs TCL, Yazar F, Thompson RM, Jurney PL. Design of a microfluidic bleeding chip to evaluate antithrombotic agents for use in COVID-19 patients. Cell Mol Bioeng 2020; 13:331-9. doi: 10.1007/s12195-020-00644-x [Crossref] [ Google Scholar]
- Qiang L, Guo J, Han Y, Jiang J, Su X, Liu H. A novel anti Candida albicans drug screening system based on high-throughput microfluidic chips. Sci Rep 2019; 9:8087. doi: 10.1038/s41598-019-44298-w [Crossref] [ Google Scholar]
- Nam H, Funamoto K, Jeon JS. Cancer cell migration and cancer drug screening in oxygen tension gradient chip. Biomicrofluidics 2020; 14:044107. doi: 10.1063/5.0011216 [Crossref] [ Google Scholar]
- Horowitz LF, Rodriguez AD, Au-Yeung A, Bishop KW, Barner LA, Mishra G. Microdissected "cuboids" for microfluidic drug testing of intact tissues. Lab Chip 2021; 21:122-42. doi: 10.1039/d0lc00801j [Crossref] [ Google Scholar]
- Liu X, Zheng W, Jiang X. Cell-based assays on microfluidics for drug screening. ACS Sens 2019; 4:1465-75. doi: 10.1021/acssensors.9b00479 [Crossref] [ Google Scholar]
- Schuster B, Junkin M, Kashaf SS, Romero-Calvo I, Kirby K, Matthews J. Automated microfluidic platform for dynamic and combinatorial drug screening of tumor organoids. Nat Commun 2020; 11:5271. doi: 10.1038/s41467-020-19058-4 [Crossref] [ Google Scholar]
- Radhakrishnan J, Varadaraj S, Dash SK, Sharma A, Verma RS. Organotypic cancer tissue models for drug screening: 3D constructs, bioprinting and microfluidic chips. Drug Discov Today 2020; 25:879-90. doi: 10.1016/j.drudis.2020.03.002 [Crossref] [ Google Scholar]
- Dhiman N, Kingshott P, Sumer H, Sharma CS, Rath SN. On-chip anticancer drug screening - recent progress in microfluidic platforms to address challenges in chemotherapy. Biosens Bioelectron 2019; 137:236-54. doi: 10.1016/j.bios.2019.02.070 [Crossref] [ Google Scholar]
- Lee WB, Chien CC, You HL, Kuo FC, Lee MS, Lee GB. An integrated microfluidic system for antimicrobial susceptibility testing with antibiotic combination. Lab Chip 2019; 19:2699-708. doi: 10.1039/c9lc00585d [Crossref] [ Google Scholar]
- Venugopal Menon N, Lim SB, Lim CT. Microfluidics for personalized drug screening of cancer. Curr Opin Pharmacol 2019; 48:155-61. doi: 10.1016/j.coph.2019.09.008 [Crossref] [ Google Scholar]
- Branchford BR, Ng CJ, Neeves KB, Di Paola J. Microfluidic technology as an emerging clinical tool to evaluate thrombosis and hemostasis. Thromb Res 2015; 136:13-9. doi: 10.1016/j.thromres.2015.05.012 [Crossref] [ Google Scholar]
- Colace TV, Tormoen GW, McCarty OJ, Diamond SL. Microfluidics and coagulation biology. Annu Rev Biomed Eng 2013; 15:283-303. doi: 10.1146/annurev-bioeng-071812-152406 [Crossref] [ Google Scholar]
- DeCortin ME, Brass LF, Diamond SL. Core and shell platelets of a thrombus: a new microfluidic assay to study mechanics and biochemistry. Res Pract Thromb Haemost 2020; 4:1158-66. doi: 10.1002/rth2.12405 [Crossref] [ Google Scholar]
- Luna DJ, NK RP, Mathur T, Bui J, Gadangi P, Kostousov VV. Tortuosity-powered microfluidic device for assessment of thrombosis and antithrombotic therapy in whole blood. Sci Rep 2020; 10:5742. doi: 10.1038/s41598-020-62768-4 [Crossref] [ Google Scholar]
- Mackman N, Bergmeier W, Stouffer GA, Weitz JI. Therapeutic strategies for thrombosis: new targets and approaches. Nat Rev Drug Discov 2020; 19:333-52. doi: 10.1038/s41573-020-0061-0 [Crossref] [ Google Scholar]
- Ahmed S, Zimba O, Gasparyan AY. Thrombosis in coronavirus disease 2019 (COVID-19) through the prism of Virchow's triad. Clin Rheumatol 2020; 39:2529-43. doi: 10.1007/s10067-020-05275-1 [Crossref] [ Google Scholar]
- Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov 2022; 12:31-46. doi: 10.1158/2159-8290.cd-21-1059 [Crossref] [ Google Scholar]
- Pilleron S, Sarfati D, Janssen-Heijnen M, Vignat J, Ferlay J, Bray F. Global cancer incidence in older adults, 2012 and 2035: a population-based study. Int J Cancer 2019; 144:49-58. doi: 10.1002/ijc.31664 [Crossref] [ Google Scholar]
- Zhang YB, Pan XF, Chen J, Cao A, Zhang YG, Xia L. Combined lifestyle factors, incident cancer, and cancer mortality: a systematic review and meta-analysis of prospective cohort studies. Br J Cancer 2020; 122:1085-93. doi: 10.1038/s41416-020-0741-x [Crossref] [ Google Scholar]
- Lu M, Zhan X. The crucial role of multiomic approach in cancer research and clinically relevant outcomes. EPMA J 2018; 9:77-102. doi: 10.1007/s13167-018-0128-8 [Crossref] [ Google Scholar]
- Mathur L, Ballinger M, Utharala R, Merten CA. Microfluidics as an enabling technology for personalized cancer therapy. Small 2020; 16:e1904321. doi: 10.1002/smll.201904321 [Crossref] [ Google Scholar]
- Duzagac F, Saorin G, Memeo L, Canzonieri V, Rizzolio F. Microfluidic organoids-on-a-chip: quantum leap in cancer research. Cancers (Basel) 2021; 13:737. doi: 10.3390/cancers13040737 [Crossref] [ Google Scholar]
- Wang HF, Liu Y, Wang T, Yang G, Zeng B, Zhao CX. Tumor-microenvironment-on-a-chip for evaluating nanoparticle-loaded macrophages for drug delivery. ACS Biomater Sci Eng 2020; 6:5040-50. doi: 10.1021/acsbiomaterials.0c00650 [Crossref] [ Google Scholar]
- Wang Y, Wu D, Wu G, Wu J, Lu S, Lo J. Metastasis-on-a-chip mimicking the progression of kidney cancer in the liver for predicting treatment efficacy. Theranostics 2020; 10:300-11. doi: 10.7150/thno.38736 [Crossref] [ Google Scholar]
- Ma C, Witkowski MT, Harris J, Dolgalev I, Sreeram S, Qian W. Leukemia-on-a-chip: dissecting the chemoresistance mechanisms in B cell acute lymphoblastic leukemia bone marrow niche. Sci Adv 2020; 6:eaba5536. doi: 10.1126/sciadv.aba5536 [Crossref] [ Google Scholar]
- Saha B, Mathur T, Tronolone JJ, Chokshi M, Lokhande GK, Selahi A. Human tumor microenvironment chip evaluates the consequences of platelet extravasation and combinatorial antitumor-antiplatelet therapy in ovarian cancer. Sci Adv 2021; 7:eabg5283. doi: 10.1126/sciadv.abg5283 [Crossref] [ Google Scholar]
- Shen P, Jia Y, Zhou W, Zheng W, Wu Y, Qu S. A biomimetic liver cancer on-a-chip reveals a critical role of LIPOCALIN-2 in promoting hepatocellular carcinoma progression. Acta Pharm Sin B 2023; 13:4621-37. doi: 10.1016/j.apsb.2023.04.010 [Crossref] [ Google Scholar]
- Haque MR, Wessel CR, Leary DD, Wang C, Bhushan A, Bishehsari F. Patient-derived pancreatic cancer-on-a-chip recapitulates the tumor microenvironment. Microsyst Nanoeng 2022; 8:36. doi: 10.1038/s41378-022-00370-6 [Crossref] [ Google Scholar]
- Chen Y, Gao D, Wang Y, Lin S, Jiang Y. A novel 3D breast-cancer-on-chip platform for therapeutic evaluation of drug delivery systems. Anal Chim Acta 2018; 1036:97-106. doi: 10.1016/j.aca.2018.06.038 [Crossref] [ Google Scholar]
- Khalid MA, Kim YS, Ali M, Lee BG, Cho YJ, Choi KH. A lung cancer-on-chip platform with integrated biosensors for physiological monitoring and toxicity assessment. Biochem Eng J 2020; 155:107469. doi: 10.1016/j.bej.2019.107469 [Crossref] [ Google Scholar]
- Llenas M, Paoli R, Feiner-Gracia N, Albertazzi L, Samitier J, Caballero D. Versatile vessel-on-a-chip platform for studying key features of blood vascular tumors. Bioengineering (Basel) 2021; 8:81. doi: 10.3390/bioengineering8060081 [Crossref] [ Google Scholar]
- Bratulic S, Gatto F, Nielsen J. The translational status of cancer liquid biopsies. Regen Eng Transl Med 2021; 7:312-52. doi: 10.1007/s40883-019-00141-2 [Crossref] [ Google Scholar]
- Bargahi N, Ghasemali S, Jahandar-Lashaki S, Nazari A. Recent advances for cancer detection and treatment by microfluidic technology, review and update. Biol Proced Online 2022; 24:5. doi: 10.1186/s12575-022-00166-y [Crossref] [ Google Scholar]
- Wang Y, Luo J, Liu J, Sun S, Xiong Y, Ma Y. Label-free microfluidic paper-based electrochemical aptasensor for ultrasensitive and simultaneous multiplexed detection of cancer biomarkers. Biosens Bioelectron 2019; 136:84-90. doi: 10.1016/j.bios.2019.04.032 [Crossref] [ Google Scholar]
- Tayoun T, Faugeroux V, Oulhen M, Aberlenc A, Pawlikowska P, Farace F. CTC-derived models: a window into the seeding capacity of circulating tumor cells (CTCs). Cells 2019; 8:1145. doi: 10.3390/cells8101145 [Crossref] [ Google Scholar]
- Tellez-Gabriel M, Cochonneau D, Cadé M, Jubellin C, Heymann MF, Heymann D. Circulating tumor cell-derived pre-clinical models for personalized medicine. Cancers (Basel) 2018; 11:19. doi: 10.3390/cancers11010019 [Crossref] [ Google Scholar]
- Xu X, Jiang Z, Wang J, Ren Y, Wu A. Microfluidic applications on circulating tumor cell isolation and biomimicking of cancer metastasis. Electrophoresis 2020; 41:933-51. doi: 10.1002/elps.201900402 [Crossref] [ Google Scholar]
- Xu M, Zhao H, Chen J, Liu W, Li E, Wang Q. An integrated microfluidic chip and its clinical application for circulating tumor cell isolation and single-cell analysis. Cytometry A 2020; 97:46-53. doi: 10.1002/cyto.a.23902 [Crossref] [ Google Scholar]
- Moghimi N, Hosseini SA, Poudineh M, Kohandel M. Recent advances on cancer-on-chip models: development of 3D tumors and tumor microenvironment. Bioprinting 2022; 28:e00238. doi: 10.1016/j.bprint.2022.e00238 [Crossref] [ Google Scholar]
- Regmi S, Poudel C, Adhikari R, Luo KQ. Applications of microfluidics and organ-on-a-chip in cancer research. Biosensors (Basel) 2022; 12:459. doi: 10.3390/bios12070459 [Crossref] [ Google Scholar]
- Xu H, Liu X, Le W. Recent advances in microfluidic models for cancer metastasis research. TrAC Trends Anal Chem 2018; 105:1-6. doi: 10.1016/j.trac.2018.04.007 [Crossref] [ Google Scholar]
- Kühlbach C, da Luz S, Baganz F, Hass VC, Mueller MM. A microfluidic system for the investigation of tumor cell extravasation. Bioengineering (Basel) 2018; 5:40. doi: 10.3390/bioengineering5020040 [Crossref] [ Google Scholar]
- Liu T, Li C, Li H, Zeng S, Qin J, Lin B. A microfluidic device for characterizing the invasion of cancer cells in 3D matrix. Electrophoresis 2009; 30:4285-91. doi: 10.1002/elps.200900289 [Crossref] [ Google Scholar]
- Tachikawa S, Kaneda S, Kumemura M, Sato R, Tsukamoto T, Fujii T. Microfluidic device fabricated by three-dimensional lithography for observation of cancer cell invasion process. Electr Eng Jpn 2019; 207:55-61. doi: 10.1002/eej.23190 [Crossref] [ Google Scholar]
- Nashimoto Y, Okada R, Hanada S, Arima Y, Nishiyama K, Miura T. Vascularized cancer on a chip: the effect of perfusion on growth and drug delivery of tumor spheroid. Biomaterials 2020; 229:119547. doi: 10.1016/j.biomaterials.2019.119547 [Crossref] [ Google Scholar]
- Ma D, Wang R, Chen S, Luo T, Chow YT, Sun D. Microfluidic platform for probing cancer cells migration property under periodic mechanical confinement. Biomicrofluidics 2018; 12:024118. doi: 10.1063/1.5030135 [Crossref] [ Google Scholar]
- Ashammakhi NA, Elzagheid A. Organ-on-a-chip: new tool for personalized medicine. J Craniofac Surg 2018; 29:823-4. doi: 10.1097/scs.0000000000004604 [Crossref] [ Google Scholar]
- Skardal A, Shupe T, Atala A. Body-on-a-chip: regenerative medicine for personalized medicine. In: Atala A, Lanza R, Mikos AG, Nerem R, eds. Principles of Regenerative Medicine. 3rd ed. Boston: Academic Press; 2019. p. 769-86. 10.1016/b978-0-12-809880-6.00044-8.
- Ashley EA. Towards precision medicine. Nat Rev Genet 2016; 17:507-22. doi: 10.1038/nrg.2016.86 [Crossref] [ Google Scholar]
- Mughal S, López‐Muñoz GA, Fernández‐Costa JM, Cortés‐Reséndiz A, De Chiara F, Ramón‐Azcón J. Organs‐on‐chips: trends and challenges in advanced systems integration. Adv Mater Interfaces 2022; 9:2201618. doi: 10.1002/admi.202201618 [Crossref] [ Google Scholar]
- Shrestha J, Razavi Bazaz S, Aboulkheyr Es H, Yaghobian Azari D, Thierry B, Ebrahimi Warkiani M. Lung-on-a-chip: the future of respiratory disease models and pharmacological studies. Crit Rev Biotechnol 2020; 40:213-30. doi: 10.1080/07388551.2019.1710458 [Crossref] [ Google Scholar]
- Kankala RK, Wang SB, Chen AZ. Microengineered organ-on-a-chip platforms towards personalized medicine. Curr Pharm Des 2018; 24:5354-66. doi: 10.2174/1381612825666190222143542 [Crossref] [ Google Scholar]
- Huh D. A human breathing lung-on-a-chip. Ann Am Thorac Soc 2015; 12:S42-4. doi: 10.1513/AnnalsATS.201410-442MG [Crossref] [ Google Scholar]
- Abulaiti M, Yalikun Y, Murata K, Sato A, Sami MM, Sasaki Y. Establishment of a heart-on-a-chip microdevice based on human iPS cells for the evaluation of human heart tissue function. Sci Rep 2020; 10:19201. doi: 10.1038/s41598-020-76062-w [Crossref] [ Google Scholar]
- Zhang M, Xu C, Jiang L, Qin J. A 3D human lung-on-a-chip model for nanotoxicity testing. Toxicol Res (Camb) 2018; 7:1048-60. doi: 10.1039/c8tx00156a [Crossref] [ Google Scholar]
- Meng Q, Wang Y, Li Y, Shen C. Hydrogel microfluidic-based liver-on-a-chip: mimicking the mass transfer and structural features of liver. Biotechnol Bioeng 2021; 118:612-21. doi: 10.1002/bit.27589 [Crossref] [ Google Scholar]
- Theobald J, Ghanem A, Wallisch P, Banaeiyan AA, Andrade-Navarro MA, Taškova K. Liver-kidney-on-chip to study toxicity of drug metabolites. ACS Biomater Sci Eng 2018; 4:78-89. doi: 10.1021/acsbiomaterials.7b00417 [Crossref] [ Google Scholar]
- Nguyen VVT, Ye S, Gkouzioti V, van Wolferen ME, Yengej FY, Melkert D. A human kidney and liver organoid-based multi-organ-on-a-chip model to study the therapeutic effects and biodistribution of mesenchymal stromal cell-derived extracellular vesicles. J Extracell Vesicles 2022; 11:e12280. doi: 10.1002/jev2.12280 [Crossref] [ Google Scholar]
- Ferreira DA, Conde JP, Rothbauer M, Ertl P, Granja PL, Oliveira C. Bioinspired human stomach-on-a-chip with in vivo like function and architecture. Lab Chip 2023; 23:495-510. doi: 10.1039/d2lc01132h [Crossref] [ Google Scholar]
- Huh DD. A human breathing lung-on-a-chip. Ann Am Thorac Soc 2015; 12 Suppl 1:S42-4. doi: 10.1513/AnnalsATS.201410-442MG [Crossref] [ Google Scholar]
- Jia L, Han F, Yang H, Turnbull G, Wang J, Clarke J. Microfluidic fabrication of biomimetic helical hydrogel microfibers for blood-vessel-on-a-chip applications. Adv Healthc Mater 2019; 8:e1900435. doi: 10.1002/adhm.201900435 [Crossref] [ Google Scholar]
- Zhao Y, Rafatian N, Wang EY, Feric NT, Lai BF, Knee-Walden EJ. Engineering microenvironment for human cardiac tissue assembly in heart-on-a-chip platform. Matrix Biol 2020; 85-86:189-204. doi: 10.1016/j.matbio.2019.04.001 [Crossref] [ Google Scholar]
- Wang Y, Wang L, Guo Y, Zhu Y, Qin J. Engineering stem cell-derived 3D brain organoids in a perfusable organ-on-a-chip system. RSC Adv 2018; 8:1677-85. doi: 10.1039/c7ra11714k [Crossref] [ Google Scholar]
- Musah S, Mammoto A, Ferrante TC, Jeanty SSF, Hirano-Kobayashi M, Mammoto T. Mature induced-pluripotent-stem-cell-derived human podocytes reconstitute kidney glomerular-capillary-wall function on a chip. Nat Biomed Eng 2017; 1:0069. doi: 10.1038/s41551-017-0069 [Crossref] [ Google Scholar]
- Özkan A, Stolley D, Cressman EN, McMillin M, DeMorrow S, Yankeelov TE. The influence of chronic liver diseases on hepatic vasculature: a liver-on-a-chip review. Micromachines (Basel) 2020; 11:487. doi: 10.3390/mi11050487 [Crossref] [ Google Scholar]
- Asrani SK, Devarbhavi H, Eaton J, Kamath PS. Burden of liver diseases in the world. J Hepatol 2019; 70:151-71. doi: 10.1016/j.jhep.2018.09.014 [Crossref] [ Google Scholar]
- Hassan S, Sebastian S, Maharjan S, Lesha A, Carpenter AM, Liu X. Liver-on-a-chip models of fatty liver disease. Hepatology 2020; 71:733-40. doi: 10.1002/hep.31106 [Crossref] [ Google Scholar]
- Gaudriault P, Fassini D, Homs-Corbera A. Heart-on-a-chip. In: Hoeng J, Bovard D, Peitsch MC, eds. Organ-on-a-Chip. Academic Press; 2020. p. 255-93. 10.1016/b978-0-12-817202-5.00008-5.
- Sidorov VY, Samson PC, Sidorova TN, Davidson JM, Lim CC, Wikswo JP. I-Wire Heart-on-a-Chip I: three-dimensional cardiac tissue constructs for physiology and pharmacology. Acta Biomater 2017; 48:68-78. doi: 10.1016/j.actbio.2016.11.009 [Crossref] [ Google Scholar]
- Liu H, Bolonduro OA, Hu N, Ju J, Rao AA, Duffy BM. Heart-on-a-chip model with integrated extra- and intracellular bioelectronics for monitoring cardiac electrophysiology under acute hypoxia. Nano Lett 2020; 20:2585-93. doi: 10.1021/acs.nanolett.0c00076 [Crossref] [ Google Scholar]
- Bang S, Jeong S, Choi N, Kim HN. Brain-on-a-chip: a history of development and future perspective. Biomicrofluidics 2019; 13:051301. doi: 10.1063/1.5120555 [Crossref] [ Google Scholar]
- Dauth S, Maoz BM, Sheehy SP, Hemphill MA, Murty T, Macedonia MK. Neurons derived from different brain regions are inherently different in vitro: a novel multiregional brain-on-a-chip. J Neurophysiol 2017; 117:1320-41. doi: 10.1152/jn.00575.2016 [Crossref] [ Google Scholar]
- Park J, Wetzel I, Marriott I, Dréau D, D'Avanzo C, Kim DY. A 3D human triculture system modeling neurodegeneration and neuroinflammation in Alzheimer's disease. Nat Neurosci 2018; 21:941-51. doi: 10.1038/s41593-018-0175-4 [Crossref] [ Google Scholar]
- Samson AJ, Robertson G, Zagnoni M, Connolly CN. Neuronal networks provide rapid neuroprotection against spreading toxicity. Sci Rep 2016; 6:33746. doi: 10.1038/srep33746 [Crossref] [ Google Scholar]
- Phan DT, Bender RHF, Andrejecsk JW, Sobrino A, Hachey SJ, George SC. Blood-brain barrier-on-a-chip: microphysiological systems that capture the complexity of the blood-central nervous system interface. Exp Biol Med (Maywood) 2017; 242:1669-78. doi: 10.1177/1535370217694100 [Crossref] [ Google Scholar]
- Peng B, Hao S, Tong Z, Bai H, Pan S, Lim KL. Blood-brain barrier (BBB)-on-a-chip: a promising breakthrough in brain disease research. Lab Chip 2022; 22:3579-602. doi: 10.1039/d2lc00305h [Crossref] [ Google Scholar]
- Yu F, Kumar NDS, Foo LC, Ng SH, Hunziker W, Choudhury D. A pump-free tricellular blood-brain barrier on-a-chip model to understand barrier property and evaluate drug response. Biotechnol Bioeng 2020; 117:1127-36. doi: 10.1002/bit.27260 [Crossref] [ Google Scholar]
- Ibrahim M, Chakrabarty K. Error recovery in digital microfluidics for personalized medicine. In: 2015 Design, Automation & Test in Europe Conference & Exhibition (DATE). Grenoble: IEEE; 2015. 10.7873/date.2015.1126.
- Briones J, Espulgar W, Koyama S, Takamatsu H, Tamiya E, Saito M. The future of microfluidics in immune checkpoint blockade. Cancer Gene Ther 2021; 28:895-910. doi: 10.1038/s41417-020-00248-7 [Crossref] [ Google Scholar]
- Arshavsky-Graham S, Segal E. Lab-on-a-chip devices for point-of-care medical diagnostics. Adv Biochem Eng Biotechnol 2022; 179:247-65. doi: 10.1007/10_2020_127 [Crossref] [ Google Scholar]