Bioimpacts. 14(3):29945.
doi: 10.34172/bi.2023.29945
Review
Regulation of cell fate by cell imprinting approach in vitro
Farkhonde Hasannejad Conceptualization, Investigation, Writing – original draft, Writing – review & editing, 1, 2 
Leila Montazeri Investigation, Writing – review & editing, 3
João F Mano Investigation, Supervision, 4, * 
Shahin Bonakdar Conceptualization, Investigation, Supervision, Writing – review & editing, 5, * 
Ahmad Fazilat Writing – review & editing, 2
Author information:
1Department of Tissue Engineering and Applied Cell Sciences, School of Medicine, Semnan University of Medical Science, Semnan, Iran
2Genetic Department, Breast Cancer Research Center, Motamed Cancer Institute, ACECR, Tehran, Iran
3Department of Cell Engineering, Cell Science Research Center, Royan Institute for Stem Cell Biology and Technology, ACECR, Tehran, Iran
4Department of Chemistry, CICECO - Aveiro Institute of Materials, University of Aveiro, Portugal
5National Cell Bank Department, Pasteur Institute of Iran, Tehran, Iran
Abstract
Cell culture-based technologies are widely utilized in various domains such as drug evaluation, toxicity assessment, vaccine and biopharmaceutical development, reproductive technology, and regenerative medicine. It has been demonstrated that pre-adsorption of extracellular matrix (ECM) proteins including collagen, laminin and fibronectin provide more degrees of support for cell adhesion. The purpose of cell imprinting is to imitate the natural topography of cell membranes by gels or polymers to create a reliable environment for the regulation of cell function. The results of recent studies show that cell imprinting is a tool to guide the behavior of cultured cells by controlling their adhesive interactions with surfaces. Therefore, in this review we aim to compare different cell cultures with the imprinting method and discuss different cell imprinting applications in regenerative medicine, personalized medicine, disease modeling, and cell therapy.
Keywords: Cell imprinting, Molecular imprinting, Disease modeling, Personalized medicine, Cell therapy, Regenerative medicine
Copyright and License Information
© 2024 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
Tissues are comprised of cells and their local surroundings, including the extracellular matrix (ECM). The ECM consists of different proteins in a three-dimensional (3D) structure with biophysical and biochemical signaling that is specific to each type of cell. These signals play a key role in diverse cell functions including attachment, proliferation, migration, expression and differentiation.1,2 For cell-based experiments involved in research or clinics, an artificial ECM is required to support cell function, which can be considered as a substrate. Cells are cultivated in a substrate sense and respond to the physical/chemical properties of the contact surface. The chemical interaction of cultured cells with substrates is gradually affected by the presence of small molecules, ions and proteins in media. On the other hand, the physical characteristics of a surface are characterized by hydrophobicity, charge, topography and viscoelasticity.3,4 The consequences of the substrates’ physical/mechanical properties on cell proliferation and differentiation have been extensively researched.5,6
There are different methods employed for 2D or 3D cell cultures in vitro. Monolayer cell cultivation in laboratories commonly relies on conventional polystyrene plates. Nevertheless, it is important to note that the physical and chemical characteristics of these 2D substrates diverge significantly from those found in natural environments. In such a way that cell-cell and cell-extracellular environment communications are not demonstrated in 2D conditions as they would be in tumor aggregates. These interactions manage cell differentiation, proliferation, cellular survival, expression of genes and proteins reaction to stimuli, drug metabolism, paracrine effects and other cellular responses. For example, the association of β1-integrin and epidermal growth factor receptor signaling was recognized in 3D culture but not seen in 2D.7,8 Therefore, it is proposed that 3D cultures can be utilized to bridge the gap between 2D and in vivo conditions in various areas such as cancer research, drug discovery, neuroscience, and regenerative medicine.
The methods for 3D culture can be classified as either scaffold-based or non-scaffold-based. The scaffold-based methods involve the use of hydrogels, fibers, or sponge-like structures made from natural or synthetic biomaterials. These structures are created using techniques such as bio-printing, freeze-drying, or electrospinning. In 3D non-scaffold-based methods, cells will be cultured in the form of spheroids, organoids, or pellets using low attachment flasks, microfluidic systems or bioreactors.8-10
Recently, the cell imprinting method has been suggested as a high throughput and cost-effective strategy for manufacturing pseudo-3D cell culture substrates. In this strategy, settled cells are utilized as formats to replicate negative designs of the cells’ membranes. These particular bio-geometrical properties are utilized as physical signals to balance cell reactions such as cell migration, adhesion, proliferation, and differentiation.11 Utilizing cells for cell-imprint makes micro-/nano structures with dynamic areas which can be engaged in within the cell–surface intuitively. Given the significance of improving the cell culture, in this review we intend to discuss the interaction between cells and various physicochemical attributes of culture substrates in 2D, 3D, and cell imprinting.
Cell culture methods in vitro
An in vitro cell culture is a procedure utilized to investigate the behavior of animal cells in a regulated environment that imitates nature without ideally changing cell morphology and function. Presently, there are various types of cell cultures. Animal cell cultures have been used to investigate fundamental cell biology, interactions of drugs or materials with cells, and the generation of vaccines and proteins, etc. Amphibian cells were selected as the primary model for the cell culture because cells in exothermic animals do not require consecutive incubation periods. Subsequently, advances in biology increased focus on endothermic animals, where normal and pathological expansion is the same as that of humans. The availability of genetically pure mouse strains facilitated the investigation of mammals in laboratory settings.12 Because the culture medium is the most important factor in cell culture technology, researchers must evaluate different cell culture procedures to choose a suitable medium or modify previous ones to suit their purposes. Artificial media can be classified into several groups based on the type of supplemented materials: serum-containing media, serum-free media, protein-free media, and defined chemical media.13
In order to conduct laboratory investigations, researchers frequently employ a variety of procedures in separation, fractionation, and characterization to prepare tissue samples accordingly. The selection of each method depends on the favorable level of separation, maintenance of viability, and technical investigation to be evaluated. Generally, highly efficient cell separation techniques rely on various factors such as cell size, cell density, cell charge, cell surface chemistry, cellular complexity and fluorescence effusion of two or more cellular components or adsorbed antibodies.12
In the next step after isolation, the cell cultures can be grown in a suspension form in the medium or allowed to adhere to a glass or plastic dish depending on the nature of the cells and their microenvironment.14 The most frequently utilized type of cell culture is the two-dimensional (2D) model; however, recently 3D culture procedures have increased in popularity.15,16
Tissue culture plates
In 2D cultures, cells adhere to polystyrene-based surfaces and are expanded as a monolayer in a culture flask or flat Petri dish.17 The principal advantage of this method is its simplicity in using a group of cloned cells. Cell cultures require significant control of the physicochemical environment (i.e., pH, temperature, osmotic pressure, oxygen, CO2 tension and chemicals), which must be regulated accurately to obtain reproducible results.12 While 2D cultures on smooth rigid polystyrene-based culture dishes are the most prevalent types of culture, mammalian tissues are predominantly pliable and textured structures. It is well known that both rigidity and roughness affect cell function in vitro.18
Therefore, to better mimic the natural environment in vitro, factors such as forced polarity, flattened cell shape, mechanical/biochemical signals, and subsequent cell-to-cell communication must be regulated.19,20 Otherwise, the 2D monolayer environment changes the function of the cells and disturbs the final outcome. For example, Proper assessment of significant properties of cancer cells is not achievable in 2D cultures.21 Hence, the main disadvantage of the 2D environment is its inability to mimic the physical mechanical properties of the cell ECM. In addition, in the 2D environment, cell-cell and cell-extracellular interactions are not the same as in the tumor environment. These communications can cause cell proliferation, differentiation and gene or protein expressions in response to chemical stimuli such as drugs or small molecules.22-24 Isolation of cells from their natural 3D environment and subsequent cultivation in vitro on 2D substrates changes the original morphology of the nucleus and cytoplasm, and affects cell function in response to signals.25,26 As a result, after several cell divisions, the cells lose their polarity during coherent expansion due to the presence of aberrations in the DNA spatial structure, which leads to the reaction of those cells to apoptosis and other phenomena.27,28 A further disadvantage of the 2D culture is that the monolayer cells have inadequate access to the essential components of the medium, including oxygen, nutrients, metabolites and signal molecules.29 Therefore, there are no similarities between the microenvironment of a cell niche and the case of an in vitro monolayer culture. In the case of tumors where several types of cells are involved, there are noted alterations in gene expression, splicing, topology, and cell biochemistry in the 2D culture.30,31
These disadvantages of 2D cultures encouraged researchers to find another model that could more aptly mimic the natural environment of cells in tissues including 3D culture systems.32
Organoid, pellet, spheroid, 3D scaffolds and hydrogels
An appropriate gradient for nutrients and waste is required to enable 3D culture media for supporting cell growth that most closely resembles the in vivo environment. The invention of 3D cell culture methods that mimic the in vivo interactions of tissues and organs has created new opportunities for researchers to investigate the biochemical and biomechanical signals during cell development.21,33 Here, we discuss the various 3D cell culture techniques such as organoids, pellets, spheroids, 3D scaffolds or hydrogels that have been developed to simulate in vivo conditions for cell growth.
Organoids are 3D multicellular structures cultivated in vitro that replicate the characteristics of an organ. They serve as a valuable tool for studying specific aspects of an organ within a controlled laboratory setting.34 Organoid cultures are potent methods progressively used for various investigations including regenerative medicine, disease modeling, drug discovery and personalized medicine. Organoids can be obtained either from pluripotent stem cells (PSCs) or adult tissue-resident cells, whether stem or differentiated cells and embryonic progenitors.35 An organoid is considered to be a 3D construct developed from stem cells including organ-specific cell types that are being self-organized via cell sorting and spatially restricted lineage commitment.36 Organoid models are more useful than other cellular models, especially in the heterogeneities of cancer, hereditary genetic disorders (such as cystic fibrosis) and host–pathogen interplays.37,38 It has been shown that organoids contribute as great tools in studying tumorigenesis and cancer development in vitro and represent a great potential for translational studies.39,40 Long ago, the approach of tumoroids, a promising elective culture show to the routine 2D system, has been driven to critical advances in understanding complex cancer cell science. The term “tumoroid” implies “tumor-like organoid”: tumoroids regularly evolve from primary tumors gathered from oncological patients.41 Cancer physiopathology can further be elucidated through specific investigations on some major aspects of characterizing the tumor microenvironment (TME). Moreover, the pathways included in the initiation of cancer and cell-to-cell connection, as well as the role of TME in cancer development are yet remained unexplained; hence, tumoroids may play a vital role in cancer biology.42,43 Upon replacement with the supporting matrix, cells inside the tumoroids can partially, imitate the in-vivo-like conditions in which morphology and cellular interactions closely resemble the primary niche. These appearances result in acquiring useful information that mimics to the human body. Tumoroids have been employed to realize if the healthy mammary microenvironment can induce a physiological action in breast cancer cells, and the reaction of cancer cells to new drugs.42 To better explore from this viewpoint, tumoroids have been made using breast cancer cells and ECM. These components have the potential to induce enormous differentiation and growth of stem cells derived from other organs after their injection into the mammary glands of mice.44 In another study, the tumoricidal properties of melatonin on colon cancer tumoroids in three different cell lines including HT29 adenocarcinoma cells, fetal foreskin HFFF2 fibroblasts and HUVECs (Human umbilical vein endothelial cells) to emulate in vivo conditions were evaluated in vitro. It was stated that the 3D tumoroid system is a promising approach for the assessment of anti-cancer compounds as compared to the conventional 2D culture system and melatonin was able to significantly decrease the cohesion of tumor mass through the arrest of some angiogenesis agents such as endocan with simultaneous induction of necrotic variations.45
However, there is limited use of organoid methods for complex and advanced diseases that are not connected to tissue growth or monogenetic drivers. In such cases, distinct phenotypes such as those in many polygenic autoimmune conditions, may not indicate the originating cell status or relevant environmental signal in an organoid method.46 Through a study on liver organoids, Huch et al reported that cells may not reach full maturity, presumably because common culturing procedures do not recreate the body’s complete biochemical and physical environment.47 For instance, organoids located outside the body of an animal do not experience natural directional signals that accurately arrange the body’s cells from top to bottom or left to right.48 A major drawback is the inability of researchers to create organs that have blood vessels. The organoid's size and complexity are constrained due to the absence of blood circulation.49 Therefore, it is necessary to consider systematic approaches to determine the advantages and disadvantages of the organoid model.
Another model is the pellet culture of the chondrocyte in vitro. It is well known that the original phenotype of chondrocytes changes to a fibroblast-like phenotype after isolation from the body and subsequent cultivation in vitro.
The pellet culture is a constant, biomaterial-free culture system that is well-suited for both chondrocyte redifferentiation and mesenchymal stem cell (MSC) differentiation studies in vitro. The pellet culture exhibits a significant cellular density, promoting extensive cell-cell contacts.50,51 Traditionally, the word pellet has been applied to describe consistently constructed geometrically determined agglomerates created from diverse beginning materials by the use of various processing conditions. Pellets for pharmaceutical purposes are routinely manufactured in a size range from 0.5 to 1.5 mm. The pellet culture provides a similar environment to the natural cartilage (dense cells in avascular hydrogel) and prevents chondrocytes from dedifferentiation.52 In addition, this technique is utilized for chondrogenic differentiation of stem cells where a number of cells (2 to 4 ×105) are centrifuged and cultured in a specified culture medium that predominantly contains transforming growth factor β (TGF-β). After 14 days, the cells form a brown nodule-like structure that expresses hyaline cartilage-specific markers such as collagen type II.53
Spheroids are developed from a simple cluster of cells such as embryonic bodies or tumors by 3D culture methods.54 Scaffold-free 3D cell spheroids can be obtained from suspension by the floating procedure, suspending drop method or agitation-based methods. Spheroids regenerate the physiological properties of tissues and tumors regarding cell-cell attachment and allow for natural cell-matrix interactions. Such structures can be produced in large numbers and be included in high-throughput screening methodologies.55,56 Spheroids can be developed to a size where oxygen and nutrient gradients are similar to the natural tissue.57 Spheroids with a radius rate of 200 µm or greater consist of an active layer of proliferating cells on the exterior and quiescent cells on the interior due to nutrient and oxygen carrier restrictions. Remarkably, larger spheroids include necrotic cells at the center which are similar to some solid tumors in vivo. Spheroids have been used to assess drug sensitivity and resistance and they are usually more durable to chemo-and radiotherapies in contrast to cells cultured as 2D monolayers.58 However, challenges exist when using spheroid models for drug delivery investigations including their formation and holding on identical size, developing spheroids from small number of cells, manufacturing tissue-like spheroids with numerous cell types, evaluating tissue-like spheroids and producing them as suitable models in drug delivery and efficacy testing.59 Furthermore, a dissemination gradient along with an enhanced spheroid size and absence of nutrients in the core of the spheroid has been observed.60 Cells in the necrotic core lose their functions due to lack of nutrition and accumulation of toxic wastes.61 These characteristics of spheroids limit drug diffusion to an extent comparable to organs and tissues in vivo.62
The use of 3D porous scaffolds has been suggested as a means of replicating the natural properties of the extracellular matrix (ECM) in tissue engineering. Balancing the attachment of cells to the scaffold with the degradation rates of the scaffolds is a key focus in enhancing tissue formation.63 Hence, a perfect scaffold should have well-determined and reproducible properties so that cells can penetrate, proliferate and differentiate inside the pores.64,65 One drawback of using popular 3D scaffolds (such as PLGA-based scaffolds) is that the hydrophobic nature of these materials prevents nutrient diffusion. On the other hand, the pore structure of hydrophilic scaffolds (collagen-based) can be filled via swelling in the culture medium prior to the cells’ penetration. Therefore, encapsulation of cells in hydrogels appears to be reasonable.
Hydrogel-based scaffolds have attracted attention because of their optimal composition and structural similarities to natural ECM and their favorable framework for cellular proliferation and durability.66 Hydrogels are 3D systems that consist of hydrophilic polymers crosslinked via covalent bonds or maintained together via physical and intermolecular attractions. Hydrogels can accommodate large contents of water or biological fluids and swell without dissolving. The high hydrophilicity of hydrogels is exclusive because of the presence of hydrophilic moieties such as carboxyl, amide, amino, and hydroxyl groups dispersed along the backbone of polymeric chains. Under swollen conditions, hydrogels are soft and rubbery, which is similar to living tissues.66,67 One of the main properties of hydrogel organization is that the biomaterials applied should be capable of self-assembly from a liquid monomeric phase to a solid polymeric mesh network.68 A hydrogel creates a humid environment that supports cell migration and attracts several exudates.69 Hydrogels suffer from a significant limitation in their mechanical strength, prompting the suggestion of chemical and molecular design approaches to enhance their toughness and strength.70
El-Sherbiny et al reported challenges dealing with hydrogels including inappropriate cell penetration and uncontrolled cell seeding due to the lack of spatial and temporal control. Several problems are related to the complexity of natural tissues comprising numerous cell types and unique ECM. In contrast, engineered tissues are composed of a single type of cell whose artificial ECM has lower mechanical properties as well as different physical characteristics at both the macroscopic and microscopic levels. This limits their use to soft and non-load-bearing tissues and the absence of intricate microvasculature in most engineered tissues which significantly diminishes the viability and function of seeded cells because of the lack of nutrient transportation and signaling molecules.66 Encapsulation of cells in alginate is widely used in laboratories due to its low cost, transparency, biocompatibility and simplicity of cell encapsulation and hydrogel decomposition. Cells suspended in their medium can be mixed with alginate solutions and encapsulated due to ionic substitutions.71 For example, a solution of sodium alginate can be added dropwise to a calcium chloride solution to form capsules. In addition, the calcium ions can be chelated to decompose the capsules and release the cells. The main problem with hydrogels is that these structures are mostly composed of a greater than 95% liquid phase, which means that cells have no place for attachment and proliferation. In addition, sample preparation of cells embedded with hydrogel exhibits more technical challenges. Techniques utilized on tissue samples, including mechanical and/or enzymatic disruption, can take advantage through liberating embedded cells but must be performed with utmost care to preserve the integrity of intracellular components while still maintaining adequate yields. The precise properties of the hydrogel must be considered as well.72 Another option is to compartmentalize cells into liquefied semi-permeable capsules, which could allow better self-assembled cellular organization and proliferation.73 In such strategies microparticles or other solid elements can be included inside these compartments as supporting elements for anchorage-dependent cells.74,75
The advancement in three dimensional (3D) bioprinting technology promotes reliability, repeatability and adaptability in the fabrication of tissue-engineered scaffolds. Bio-inks containing printable biomaterials, cells and small molecules (cell-laden biomaterials) are being utilized in bioprinter to form a 3D structure in a layer-by-layer fashion.76 In order to enhance the capacity of regeneration, bio-inks with similar characteristics to the natural environment of cells are proposed.77
Microfluidic and its application
Microfluidics is a well-accepted physics field that is currently used to expand cell biology devices.78 By downsizing microscopic systems and taking advantage of accurate processing, high-power biological experiments can be conducted on chips. Particular properties of dynamic flow at the micrometer-scale, spatial regulation of liquid constitution at subcellular resolution, rapid media and temperature variations, and single-cell management and investigation are the main pros of this method.79 The aim is not to create a whole living organ, the aim is not to create a whole living organ, but to at least manufacture operational units that carry tissues and organs fundamental roles. The elementary model is a single, perfused microfluidic chamber which includes one type of cultured cell presenting functions of a distinct tissue type. In more intricate patterns, two or more microchannels are linked by permeable membranes that are coated on opposite sides by various cell types to renovate junctions between several tissues.80,81 These systems can involve physical powers such as physiologically related levels of fluid shear stress, cyclic strain and mechanical pressure, and permit analysis of organ-specific reactions involving immune cells’ response to drugs, toxins or other environmental disturbances.81
Initially, the innovation in soft lithography and molding of polymers facilitated the development of inexpensive microfluidic tools including open microfluidic devices,82 which have further benefits because of the physical properties of these polymers.83 The most prevalent technology for constructing microfluidic devices for use in cell biology is based on the soft lithography of polydimethylsiloxane (PDMS). PDMS is an elastomer that can be built into microfluidic devices via ordinary molding methods.84 Its broad application as a suitable material is because of its flexible feature to integrate fluidic gates which are necessary components for main microfluidic use.85 Numerous applications of microfluidics in cell-based devices have been acknowledged. As another possible alternative to traditional cell culture and animal studies, human organs-on-chips or microfluidics could change many areas of basic investigation and drug development.81 Microfluidic chips regulate many system parameters that are not simply managed in 3D static cultures, and help to investigate a wide range of physiological phenomena. They could be used to explore the molecular processes of organ growth and disease and the interplay of the body with stimuli such as drugs, environmental factors, consumer supplies and medical devices.86 Microfluidic devices can be used to examine physiological and pathological mechanisms that take place within a relatively short period (approximately <1 month) and rely on comparative cell locations within organ- or tissue-specific microarchitecture.81,87 For instance, many aspects of hepatic physiology can be investigated by the use of liver-on-chip applications and include the generation of liver-specific proteins, polarized cell displacement of lipids and other species, and energy metabolism of various patient populations. Stress reactions that include an acute phase reaction as well as ischemia, partial hypoxia or nutritional exclusion can also be evaluated.88 On the other hand, the presence of fluid in the microfluidic system leads to the production of physical or chemical gradients, which have been used for noninvasive investigation of cell migration,89 cardiac tissue organization,90 nerve axon development91 and metabolic response,92 differentiation93 and neurotoxin reactions,94 in addition to the evaluation of subcellular construction95 and cell-cell junctional cohesion.96 Microfluidic systems recapitulating anatomic sites including kidney, intestine, lung, heart, smooth and striated muscle, fat, bone marrow, cornea, skin, blood vessels, nerves, and blood-brain barrier, fat and bone have been investigated over the past few decades. Many of these systems are not considered to be models of organs since only one cell type was developed in one microchannel. Nevertheless, the results of studies indicate that fluid flow and shear stress alone pose a significant impact on cell shape and function.81 Microfluidics can be used for the thorough study of absorption, distribution, metabolism, exclusion and toxicity of chemicals in vitro, instead of animal models, which is commonly the case in the pharmaceutical industry.97,98 Over the past decade, many researchers have focused on the use of microfluidics to study drug properties and confirm pharmacokinetic and pharmacodynamics modeling, and evaluate drug potency.99 This capability allows researchers to directly monitor cell behavior after drug treatment or investigate the spread of local chemical stimuli in the cell.79 For instance, a liver microfluidic system showed the interactions of different processes with drugs including drug bio-activation, drug clearance, drug-drug interactions via induction/inhibition mechanisms, sensitivity to drug-induced liver damage, and generation of reactive metabolites that could cooperate with other organs.81
Overall, microfluidics research is dealing with specific technical challenges. One disadvantage is that bubbles in microfluidic channels may damage cell membranes and prevent the proper function of chips and it is difficult to entirely remove them.100 Further problems include achieving robust, stable cell seeding in microfluidic channels, banning microbial pollution, and regulating the cell-cell and cell-ECM interactions, all of which should be resolved in order to establish precise tissue structure-function relationships.81 A fundamental issue that must be improved is the use of an appropriate material for composing microfluidics. Most microfluidics are fabricated from PDMS because of its simplicity, optical transparency, gas permeability and biocompatibility. However, the absorption of serum proteins, small organic materials such as drugs, and high gas permeability can limit the use of PDMS.101,102 Stability is another primary constraint. Different factors must align to attain the optimal role of microfluidics over a month or longer, such as cells and ECM coverings, fluidic regulation, bubble elimination and gradient preservation.103 Therefore, given the above constraints and recent successes with the use of microfluidics in imitating specific functions at the organ level, this procedure is still in its infancy. However, microfluidics are not expected to be replaced with animal experimentation in the near future because of tough regulatory requirements and complexities in recreating the roles that organs play in the body which researchers are facing. Though with the increasing number of studies about microfluidics, it is possible to gradually replace an animal-based approach at any time.81
Replicating cell shapes by cell imprinting
Cell functions are regulated by biochemical and biophysical signals that are specific to different types of tissues. This regulation is necessary because cells are situated within distinct spatial microenvironments.104 In order to understand the process of tissue reconstruction, it is necessary to consider the impact of dysregulation on tissue malfunction and how drug therapy can restore pathologically damaged tissues. This understanding requires a highly simulated environment that replicates the physicochemical factors found in the cellular microenvironment, including growth factors, ECM proteins, mechanical landscape, and topological cues.65,105,106 Based on these factors, several methods have been expanded that partially control biological interactions between cells and their surroundings.107 The extensive applications of relocating cellular features and molecular patterns on polymer surfaces have recently gained significant attention in the literatures.108,109 The presence of microscale and nanoscale topographies induces changes in the morphology of cells, affecting focal adhesion and cytoskeletal tension. Consequently, these alterations lead to modifications in nuclear shape and ultimately influence cell function.110,111 Topographic features can be engraved in microparticles that can be combined with cells that will experience such surface signals in a pseudo-3D space.112
Researchers have found that in addition to chemical recognition, the assessment of the topography of cell-imprinted substrates is also significant. Although adhesion forces mediated by molecular recognition are undoubtedly stronger, weaker binding forces resulting from topography may also become relevant after prolonged contact. It has been observed that cells show a greater inclination to attach to rough surfaces rather than smooth ones.113 Also, the specifically induced membrane forms, which are finger-printed based on the mature cell types utilized as templates, could regulate the selective activations of genes of the printed mature cells, followed by auto-activation of specific complex cell signaling and metabolomic pathways. Therefore, it seems that cell membranes, owing to their unique proteins and compounds, may play a role in the topographical pattern of the cell-imprinted substrates.114 Fig. 1 demonstrates a schematic view of the cell imprinting process and alteration in cell shapes after cultivation on the imprinted substrate.
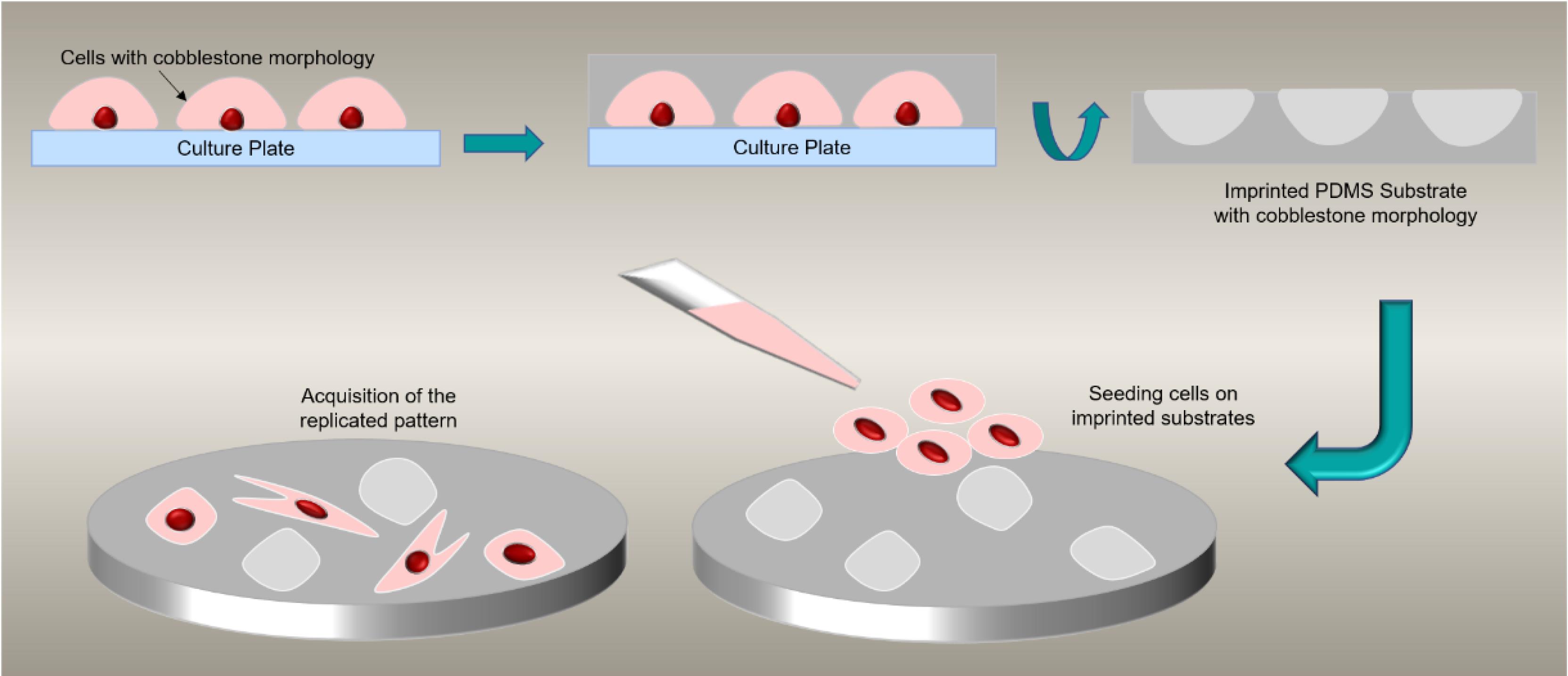
Fig. 1.
Schematic view of cell imprinting process. Seeded cells acquire cobblestone morphology on replicated shapes.
.
Schematic view of cell imprinting process. Seeded cells acquire cobblestone morphology on replicated shapes.
Generally, cells are impacted by their environment characteristics which can be explained in terms of chemistry, topography, elasticity or external stimuli. Different cell functions such as adhesion, migration, proliferation or differentiation are affected by the microenvironment and can undergo changes by processes that range from morphogenesis to cancer progression. For example, the effect of mechanical forces on a cell’s fate has been extensively investigated. It has been reported that there is a potential for matrix elasticity115 and topography to control the stem cells’ fate.116,117 Topographical patterns influence membrane and nucleus mechanics through the rearrangement of the cytoskeleton.118 According to a research, the forces applied to integrins are directly transferred (forwarded) to the nucleus, which leads to its stretching and deformation.119 Actin and other intermediate filaments interfere with the transfer of the force and microtubule stability of the nucleus.120 Since the nucleus is the largest and most rigid organelle, the tensegrity (tensional integrity) model indicates that it would be the last point affected by morphological deformation or mechanical tension to the cell.121 In addition, it has been confirmed that the nucleus, its nuclear membrane and chromatin are directly connected to the cytoskeleton via the linker of the nucleus-cytoskeleton complex (LINC), the nuclear pore complex and the underlying lamina.122 The results of recent studies prove the presence of intrinsic nuclear mechano-transduction pathways, which indicates that the structural formation of the nucleus can sense and react to mechanical tensions. Hence, the nucleus may be a mechano-sensitive structure.123-125
An important research challenge is the generation of artificial systems which can mimic the detection mechanisms occurring at the molecular level within living systems. The evolution of molecular imprinting provides a valid contribution to this research challenge. With this technology, elective molecular identification sites are presented in a polymer and can therefore mimic the biological properties of the microenvironment. Possible uses of these systems include affinity separations, medical diagnostics, drug delivery and catalysis. Recently, biosensing systems that apply molecular imprinted membranes have become the focus of research in different fields. In these systems, imprinted membranes are being utilized as biomimetic recognition components combined with a transducer element.126 According to the ‘‘lock and key’’ process which is similar to natural receptor-ligand interactions as well as easy procurement, molecular imprinting has been extensively used for almost half a century.109 The first molecular imprinting-based synthetic receptors were introduced by Wulff in 1972 and then this technique became one of the most impressive and adaptable procedures for combining particular molecular recognition sites into a polymeric network to form a receptor-like element. The molecular imprinting mechanism generally includes self-assembly of a template molecule and functional monomers through non-covalent or reversible covalent bonds in order to achieve spatial and matching replication of the form and functionality of the targeted ligand.127
Molecular imprinting
Recently, imprinted polymers (MIPs) at molecular level have become a perfect approach to the specific determination of target molecules in complex matrices where other analogous and comparable structural compounds could coexist.128 This structure particularly mimics the "lock and key" binding mechanism that occurs in the natural bio-recognition process.126,129 In a general sense, MIP as a polymeric system, contains binding cavities in a specific template. Although several fabrication techniques have been suggested over the years, all of them almost follow a similar underlying process which is summarized in Fig. 2. It is demonstrated here that the MIP is acquired by using in situ co‐polymerization of functional monomers and cross‐linkers around templates (or targets).129 During polymerization, the template is integrated into the polymeric matrix and chemical groups of functional monomers are reorganized according to the structure and chemical attributes of the template molecules. The use of the template from the acquired polymeric matrix will form a template complementary recognition site that has high substrate selectivity and specificity.126,130 After polymerization, the templates are brought out from the resulting polymeric network (e.g., by cleaning it using a dissolvent), leaving fixed holes of the native template that correspond to its form, size and orientation.129 Based on the interactivities between the functional monomer chemical groups and templates, MIPs have been categorized into covalent, non‐covalent (e.g., hydrogen bonding, van der Waals or coulomb forces, hydrophobic interactions), or semi‐covalent.131 In covalent MIPs, covalent bonds are organized between the template and functional monomers prior to polymerization; then severed during the template removal phase and reformed during rebinding. Altogether, this process produces cavities that are better defined and more uniformly distributed; however, the needed method is complicated and time-consuming, and because covalent bonds are required, there is a limited selection of monomer–template combinations.132 In the non‐covalent approach, the binding sites are organized through the self‐assembly of the monomer and template molecules in the pre-polymerization mixture and are then fixed after polymerization.133 NoncovalentMIPs are relatively simpler to prepare and the templates can be removed easier. However, noncovalent cavities may not be as uniform as covalent cavities. Semi-covalent MIPs combine two previously mentioned techniques, covalent bonds are formed between the template and functional monomer; after polymerization, the template is removed by cleaving these bonds and the analyte non-covalently attaches to its binding sites.134,135 (Fig. 2)
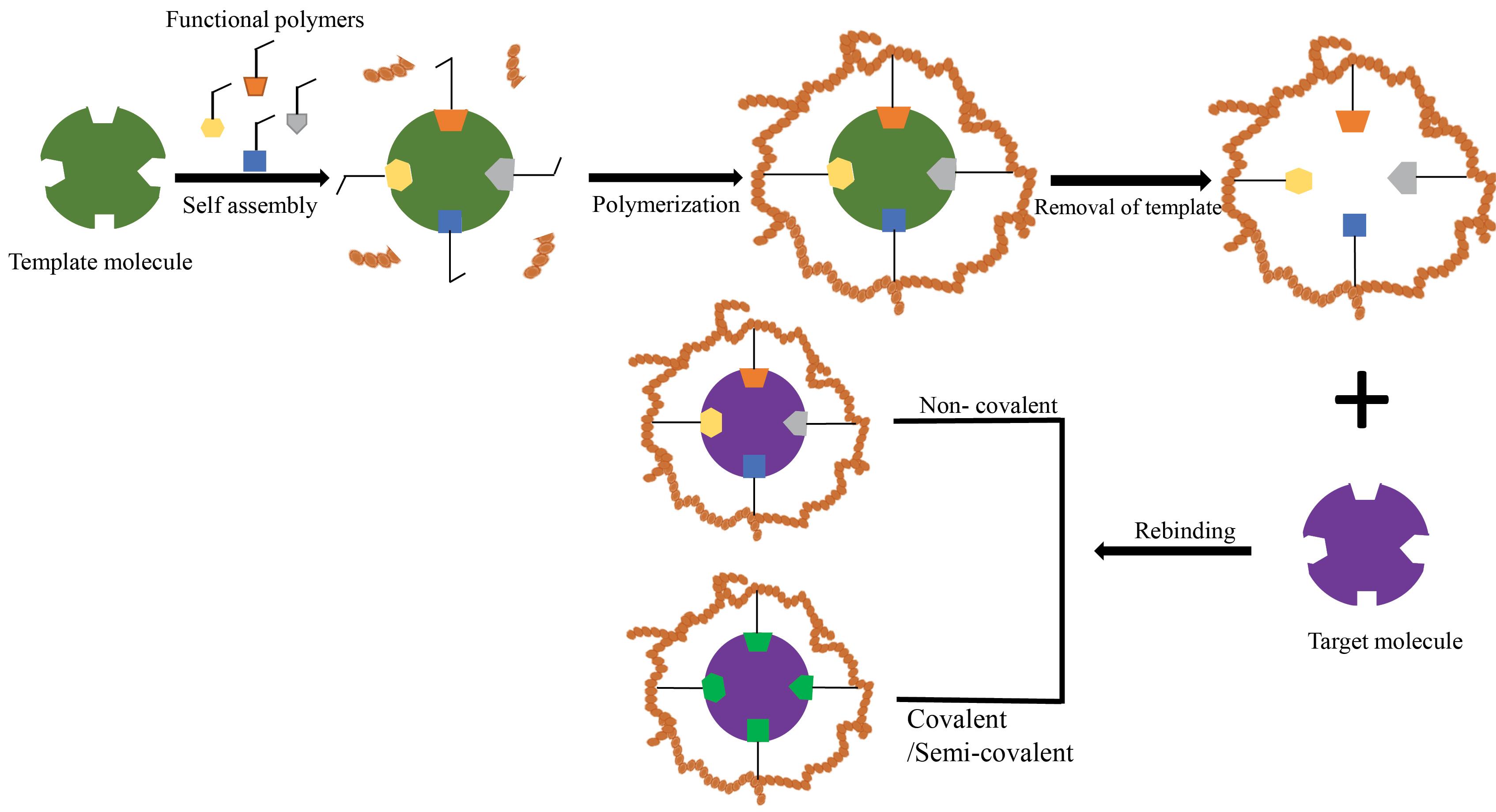
Fig. 2.
The schematic view of the MIP procedure.
.
The schematic view of the MIP procedure.
Attractive properties of MIPs include mechanical, thermal, and chemical consistency; simplicity of preparation; decreased expense; and receptor-like affinity to targeted molecules.127 MIPs are considered to be ‘‘artificial receptors’’ or ‘‘plastic antibodies’’, which can mimic the biological role of a natural receptor or antibody. Therefore they have the potential to communicate with cells and induce particular cell signaling, intracellular cascades and subsequent modulation of cell actions.
It has been found that one of the most attractive features of natural receptors is their high responsiveness to external stimuli (such as temperature, pH, etc.). Hence, in order to realize an analogy between the synthetic and the natural receptors, stimuli-responsive MIPs (SR-MIPs) have engaged substantial research attention in recent years.109 The credible and general approach to design stimuli-responsive MIPs is to exploit the molecular imprinting technique in stimuli-responsive materials, which yields the MIPs with the capacity to respond to external stimuli while regulating their affinity for the target molecules and presenting a switchable ability of the binding or releasing activities.136 Various studies have concentrated on expanding biological SR-MIPs to imitate human immune responses such as endogenous stimuli (e.g., reactive oxygen species, variations in physiological pH and temperature, overexpressed protein and enzyme levels) and exogenous stimuli (e.g., temperature, light, changes in the magnetic field, etc.). Also, the recognition ability of SR-MIPs in changes in the environment to simulate dynamic antigen-antibody or bioreceptor-ligand interplay in an organism has been investigated.136,137
However, the primary consideration in MIPs is predominantly focused on its use in adsorption, separation sensing and catalysis of molecules.127 In order to identify a cell, MIPs should have a specific binding affinity to special cell membrane molecules (e.g., proteins, lipids and glycans) or the cell membrane assembly.138,139 Therefore, approaches for MIPs that mediate cell recognition are commonly placed into two categories127: 1) cell membrane-molecular imprinting and 2) whole cell imprinting procedure. Cell membrane-molecular imprinting generally refers to the imprinting of specific parts of cell membrane molecules such as glycan chains and epitopes of membrane proteins.140,141 In addition, imprinting a bioactive ligand (e.g., proteins or peptides) that has specificity to cell membrane receptors could allow for indirect cell identification.142 However, for cell-membrane-molecular imprinting, the selection of suitable cell membrane molecules throughout the imprinting procedure is critical to the ultimate efficiency of the cell’s recognition capability.127
This procedure is appropriate for imprinting microorganisms like bacteria and viruses as they are comparatively stiff. Cell diagnosis based on the whole cell imprinting scheme relies not only on the cell-like cavities but also on the numerous non-covalent interactions between MIP matrices and cell membranes. Together, these two imprinting strategies have complimentary benefits that enable excellent feasibility and cooperation in the design of MIPs for selective cell recognition.127 The combination of the said two techniques may provide a structure with similar features at both the molecular and cellular scales.
Molecular imprinting
Nanostructured materials in MIPs
With the focus on molecular imprinted technology, MIPs with various forms and scales have been promoted so far.143 In recent years, great advances have been achieved in the production of functional nanomaterials and researchers have assigned themselves to investigate MIP nanomaterials (diameters less than 200 nm) with the same particle size and controllable morphology. Compared to conventional MIPs, MIP nanoparticles (MIP-NPs) exhibit a great dispersibility, high surface-to-volume ratio and easy rinse of template molecules, also having a high binding capacity, high selectivity, high affinity and excellent water compatibility.144 Hence, MIP-NPs applications are currently being expanded, certainly for biomedical separation and detection. This includes using them for bioimaging,145,146 diagnosing cancer,147 delivering drugs to specific targets148 and releasing drugs in a controlled way.149 MIP-NPs are anticipated to become a precious research tool for many different kinds of biomedical studies. Molecularly imprinted nanogels (MIP-NGs) are also a promising innovative material in the biomedical field. MIP-NGs as swollen hydrogel nanoparticle networks formed from either hydrophilic or amphiphilic polymer chains. They have a few key characteristics such as being stable composition, size change ability and low toxicity, stable in serum and capable of effectively encapsulating drugs. Because of these qualities, they are known as advanced systems for drug delivery. So, MIP-NGs can be very useful in disease diagnosis, delivering drugs to treat cancer and capturing images of inner parts of the body.150 Currently, MIP-NMs’ applications are extended in biomedicine since they can be selective and effective same as natural receptors. In the past few years, the molecules used to make nanomaterials are not exclusively comprised of small molecules, but also biological macromolecules (like proteins), cells different microorganisms, viruses, etc.136 For example, Zeng et al fabricated MIP-NPs with an affinity to the hydrophilic peptide GFP-9 by inverse microemulsion polymerization. The surface has a spot where peptides can stick easily and can strongly attach to the template molecule GFP-9. For imprinting short peptides, the key point is to use it as an epitope to achieve the respective macromolecular recognition.151 In another study, Teixeira et al discovered that growth factor-β3 (TGF-β3) MIP-NPs produced through epitope-imprinting reverse microemulsion polymerization, can identify and selectively attach to TGF-β3 even in complex human fluids (platelet lysate). Furthermore, adipose-derived stem cells were cultivated with MIP-NPs and incubated with platelet lysate resulting in an increased collagen II-rich matrix. This offers a cost-efficient, reliable and easily expandable substitute for non-living growth factor molecules to direct cell fate. In addition, it was confirmed that MIP-NPs can serve as GF chelating ligands to control cell activity and the operative surface of MIP-NPs can selectively identify and seize TGF-β3 and 3D cell cultures using MIP-NPs, which can enhance the accumulation of chondro-related matrix components.152
There are numerous techniques for preparing MIP-NMs for small biological molecules that are imprinted like in-site polymerization, surface imprinting, precipitation polymerization, suspension polymerization and so on. Each approach possesses distinct benefits and drawbacks like the straightforward preparation and appropriate execution of precipitation polymerization. Furthermore, the resulting MIPs exhibit uniformity in their structure and are effortlessly adjustable in terms of their size. However, the template molecules enclosed within them are not readily extractable. Furthermore, if the desired compound is specifically identified in the mixture, the remaining template molecule in the MIPs could detach, leading to potential inaccuracies in the experiment. Hence, while getting ready MIP-NMs, the particular employed approach relies on the actual circumstances.136,153
Cell imprinting
Cell imprinting is a technique that replicates cell shapes using cells as a mold to produce substrates with cavities analogous to the original cellular morphology. Different types of hydrophobic or hydrophilic materials are suggested for this process. Hydrophobic materials such as PDMS are stable at room temperature and can physically replicate microscale and nanoscale features due to the molecular structure of its backbone (e.g., Si=O). It is proposed that in hydrophilic materials such as polyacrylamide or methacrylated gelatin (GelMa), the molecular interactions between functional groups (carboxyl and amine) of materials and cells play a role in the imprinting process, which might provide an increased affinity. However, the possible evaporation/absorption of water or molecules may damage its features and interfere with the outcome. Another essential criterion for material selection is elasticity. The effects of substrate stiffness on cell expansion have been extensively evaluated.154 PDMS is well-known for its tunable stiffness and surface properties, ability to mold, optical clearness, gas permeability and lack of toxicity.155
Nevertheless, the hydrophobic surface of PDMS prevents impressive cell adhesion to the uncoated PDMS surface. The results of studies have shown that special receptors on cell membranes can sense alterations in ECM hardness. The physical characteristics of the ECM are recognized by integrins that connect ECM to the actin cytoskeleton inside the cells and transfer it to the nucleus through the LINC complex to induce nucleus rotation and reorientation. The signals are translated by changes in nucleus conformations and respond by gene or protein expressions.156-158
In a general cell imprinting procedure, cells are freshly isolated from tissues, cultured on polystyrene dishes, and fixed at the highest confluency that does not sacrifice the cell’s original phenotype. PDMS substrates are subsequently prepared by mixing a silicon base elastomer and curing agent. The fixed cells are used as a mold to make a replicate with prepared PDMS. The solid form of PDMS is separated and utilized as a new substrate for further analysis.159,160
This is a simple, safe and user-friendly process and the key element of this protocol is the preparation of cell-based molds that have the highest confluency and lowest changes in the original phenotype. This imprinted substrate could be used as a substitute for conventional tissue culture polystyrene-based dishes.161
Cells cultured on imprinted substrates
The cell membrane has intricate chemical and topographical features that are frequently identified as flexible and textured patterns. Nevertheless, the smooth and stiff transparent polystyrene (PS) tissue culture plates extensively applied for in vitro cell cultures show different physical properties as compared to ECM.162 Despite numerous endeavors to improve the connection of cells that are attached to PS plates, these substrates are orders of magnitude stiffer than soft tissues.115 The results of previous studies indicate that PDMS, at a 1:10 ratio of curing agent, induces more osteogenicity in stem cells than polystyrene plates.163,164 In some studies, engineering approaches such as lithography have been used to generate substrates with 3D features that mimic physiological shapes and patterns similar to native tissues. For example, aligned microgrooves are fabricated by photolithography to resemble the physiological dimensions of the ECM structure in tendons, muscles, nervous and cartilage tissues.165 Engineered topographical substrates which imitated natural topographies have been used to regenerate whole tissues in vitro, such as corneal substitutes166 and vascular graft.167 However, the mechanism underlying cells’ response to topography is not yet fully clear at the cellular level. Recently, there have been remarkable studies showing that topographical factors including size, shape and geometric features can have important effects on many cell behaviors such as adhesion, migration, alignment and differentiation.168-170 Among these factors, topography size seems to play an important role in modulating cell behavior. Although the cell reaction to topography is different and depends on the cell type, the effect of topographical size (such as width, distance and depth of surfaces) exists for all cell types. Whereas microtopography (such as size greater than 10 μm, which is the length scale of a mammalian cell) mostly influences entire cell morphology, nano topography is included especially with subcellular detecting instruments. The relation between the size of topography and cellular sensing organelles has been shown in various investigations. Most importantly, topographic features that have an optimal size as the most potent inducer of cellular response, have been mostly present in both the length scales. For instance, a basic surface roughness value of 1.1nm was recognized to influence the rate of proliferation for murine osteoblasts.171,172 In conditions of roughness higher than this value, the rate of cell proliferation decreased. A threshold distance of 73 nm between the nanoislands attached to the adhesive ligand was also observed to regulate the integrin clustering process and then monitor the adhesion of MC3T3 osteoblasts on the nanotopographic surface.173 At the micro length scale, human corneal epithelial cells are able to change their alignment on grooved substrate when the pitch is larger than a threshold range of 0.8–1.6 μm.174 In this manner, such data concerning the importance of topographical size would be supportive in planning for appropriate materials and platforms in tissue engineering and regenerative medicine usages.171 In Table 1, cell imprinting is compared with other cell culture methods.
Table 1.
Advantages and disadvantages of different cell culture approaches
|
Cell culture method
|
Advantages
|
Disadvantages
|
| Imprinting on a chip |
Monolayer/all cells are accessible
Safe in signaling
Wide range of applications (tissue engineering or cancer research, routine cell cultures)
Capable of producing realistic in vitro models
More reliable/predictable/repeatable
A valuable tool for personalized medicine
Induction of both physical and chemical cues
Improved cell-to-cell interactions compared with other 2D cultures 175 |
Need specific facilities to produce the chips
Lack of standardized platforms175,176 |
| Polystyrene plates |
Monolayer/all cells are accessible
Simple/priceless
Popular/standardized 177 |
Totally irrelevant to the native tissues
Induction of wrong signals to cultures
Cells lose their phenotype 178 |
| Alginate hydrogels |
3D hydrogel
Simple/priceless
Popular/standardized 179 |
Chemically irrelevant to the native tissues
Low cell attachment and proliferation due to the presence of a substantial amount of water 180 |
| Traditional imprinting |
Monolayer/all cells are accessible
Simple/priceless
Safe in signaling 181 |
Low efficiency/unpredictable/unrepeatable
Lack of standardized platforms 181 |
| Aggregate/ pellets/ organoids/ spheroids |
3D culture
More accurate representation of in vivo cell-to-cell and cell-to-ECM signaling 182 |
Difficult in cell expansion
Difficult in oxygenation
-Transiently resemble cell organization and interactions
Difficult to maintain long-term cultures 182,183 |
Scaffolds/ 3D bio-printing
|
High reproducibility
Co-culture ability
Chemical-physical gradients 184 |
Lack of vasculature
Challenges with cells/materials
Difficult for cell analysis 184 |
In addition, it has been shown that surface topography features can influence cellular drug uptake. As previously mentioned, biological science research and drug evaluation are mainly based on 2D cell culture techniques, which are unable to provide a reliable relevant physiological environment.185,186 According to studies, binding to the integrin-targeted cell-adhesive peptide RGD (Arg-Gly-Asp) segments plays a role in cell interaction with the bio-imprinted substrate of cells in vitro. It has also been discovered that cancer cells changed their β1 integrin expression, cell morphology, the organization of their cytoskeleton and proliferation when cultured on cell imprinted substrate surfaces in comparison with a 2d surface.142,187 Therefore, a suitable substrate can have significant effects on drug susceptibility, gene expression and protein synthesis by having the chemical composition, topographical and mechanical properties of the substrates.11 In a study conducted by Shahriyari et al the role of cell imprinting on viability and drug susceptibility of breast cancer cells to doxorubicin (DOX) was investigated. According to their study, the response of MCF7 cells to DOX was monitored for 24 hours. Although the biocompatibility was increased in the imprinted substrates, the cultured cells were more sensitive to the drug as compared to the plain substrates. MCF7 cells on imprinted PDMS and GelMA substrates demonstrated respectively 37% and 50% higher cell death as compared to the corresponding plain PDMS and GelMA. In addition, the cells on the imprinted hydrogel showed about 70% more drug sensitivity than the cells cultured on the imprinted PDMS substrates. Therefore, they concluded that multiscale cell membrane topography can mediate intracellular signaling and drug uptake.11 Also in another study Domura et al. stated that the morphological spreading factors (nucleus/cytoplasm area ratio) caused by the natural substrates are related closely with the cellular proliferation and the IC50 drugs of two different types of breast cancer cells (MDA-MB-231 and MCF-7).188 Tan et al. also investigated the role of paclitaxel and doxorubicin on the expression of caspase 3, the expression of proliferating nuclear antigen (PCNA), the number of cells and the secretion of vascular endothelial growth factor (VEGF) on endometrial cancer cells cultured in PDMS substrate. Based on their study, a culture substrate with cell-imprinted topography, which has nano- and micro-resolution, is able to regulate the response of endometrial cancer cells to chemotherapy drugs. Their results revealed that the topography affected the cell reactions in a drug-dependent manner. So the topography-related susceptibility of paclitaxel and doxorubicin effects varied. A culture substrate with cell-imprinted topography, which has nano- and micro-resolution, is able to regulate the response of endometrial cancer cells to chemotherapy drugs. Hence, it seems that the physical architecture of the cancer cell and the components of the nano- and micro-environment may be an appropriate prospective target to increase the clinical activity of traditional drugs. Furthermore, studies’ results imply that the cells discriminate between the various cell-like topographies (positive and negative bioimprints), suggesting that a practical topography is most desirable as a growth platforms in experiment design.189
Maintenance or restoration of the original phenotype (redifferentiation of dedifferentiated chondrocytes)
Regulation of cell function with a reliable and low-cost method is still regarded as a significant challenge. In fact, the fabrication of a substrate with simple features (grooves, ridges or pits) and basic geometries may not provide enough signal to mimic the complex structure of natural ECM. In addition, the characteristic features and design should be optimized for each cell type.190 In a similar manner, a substrate that can recreate the natural niche of the target cells can initiate particular signals driving a phenotypic change.191 For example, Mahmoudi et al. utilized cell-imprinted substrates to induce chondrogenesis, proposing that stem cells react to the design of local chondrocytes imprinted on their culture substrate.114
Although autologous chondrocyte implantation (ACI) is an approved procedure used to repair cartilage lesions, this method has several limitations such as the patient’s age (<45 years) and size of the lesion (<6 mm). In addition, ACI cannot be used for patients with osteoarthritis. During the past 20 years, numerous research endeavors were made to improve ACI procedures; however, there was less progress reported for the cellular part. Chondrocytes lose their original phenotype once isolated from the body and cultured in vitro.192 The spherical morphology of the natural chondrocytes changes to a spindle-like morphology and there are changes in their specific marker expressions, mostly in collagen II to collagen I. In order to prevent dedifferentiation, a special culture medium supplemented with TGF-β and 3D cultures such as alginate encapsulation that can maintain the chondrocyte phenotype in vitro has been proposed. TGF-β, like most growth factors, has no or limited approval for clinical applications due to the risk of side effects.193 For example, palifermin is a keratinocyte growth factor (KGF) prescribed for severe oral mucositis patients after chemotherapy. Chondrocyte imprinted substrates can maintain the original chondrocyte phenotype or restore the phenotype in dedifferentiated chondrocytes. Hence, dedifferentiated chondrocytes that have fibroblast-like morphology and collagen type I expression could re-differentiate to a spherical morphology and collagen type II expression after cultivation on a chondrocyte imprinted substrate.194
Imprinting offers several advantages over growth factors in terms of safety, stability, and reproducibility of outcomes. While growth factors are highly effective molecules, they can potentially expire or lose their functionality. In contrast, imprinting has the ability to physically generate signals that regulate cell function. However, no existing literature compares the effectiveness of growth factors to imprinted substrates. Additionally, it is important to consider the potential undesirable signals that may result from the combined use of growth factors and imprinting. Therefore, it is crucial to optimize the amount of growth factor when incorporating physical signals.195
One main problem in autologous cell transplantations is the shortage of accessible cells. Expansion of normal cells in vitro changes their original phenotype because of alterations in their environment.196 In terms of phenotype, the dedifferentiation of chondrocytes is characterized by a decrease in the expression of specific markers, including collagen type II, aggrecan, and transcription factor SOX9, and an increase in the expression of fibroblastic markers such as collagen type I and versican.197-199 The significance of this is crucial in the field of cartilage tissue engineering as dedifferentiated chondrocytes tend to transform into fibroblasts rather than producing hyaline cartilage, which is necessary for the functioning of articulating joints.199 Therefore, the loss of a chondrocyte phenotype prior to use in experimentation or implantation is of utmost importance for tissue engineering.
Induction of differentiation in stem cells
Regardless of the extensive attempts to accurately control stem cell fates with engineered patterned substrates, a trustworthy and inexpensive method that regulates stem cell behavior outside the body remains to be developed. The ECM has a complex regular structure and the construction of simple geometries that contain grooves, ridges, dots or pits cannot adequately recapitulate the ECM architecture. Designing an optimal topography for all cell types requires an extensive effort.190
It is apparent that changes in the shape of a nucleus will change the cell’s fate.158,200 The nucleus is linked to the cytoskeleton and it is presumed that alterations in nuclear topography at the nanoscale level are induced by the cell’s shape. In a recent study by Kamgouyan et al, an analysis using atomic force microscopy was performed on imprinted substrates derived from five different types of cells: osteoblasts, tenocytes, stem cells, fibroblast-like cells, and C28 cell lines. The findings of the study revealed that there were unique nanopatterns observed in the roughness parameters including amplitude, spatial, and hybrid for each individual cell type. 201 Therefore, the effect of the geometry of the nucleus on contacts between chromatin fibers and their adjustments should be investigated.122,202
Table 2 enlists published studies that pertain to cell imprinting. Researches have shown that cell imprinted substrates can induce chondrogenic,114,194 keratinogenic,158 osteogenic,200 tenogenic,203 neurogenic,204 myogenic205 and Schwan cell206 differentiation in stem cells. In addition, Abadi et al reported improvements in cardiomyocyte differentiation when induced pluripotent stem cells (iPSCs) were cultured on cardiomyocyte imprinted substrates.207
Table 2.
Summary of studies on cell imprinted substrates.
|
Year
|
Author
|
Description of the study
|
Ref.
|
| 2012 |
Jeon and Kim |
Bone mineralization of osteosarcoma (MG63) on MG63 imprinted patterns |
208
|
| 2013 |
Mahmoudi et al |
Chondrogenic differentiation of rabbit adipose-derived stem cells (ADSCs), changes in cell morphology |
114
|
| 2014 |
Mashinchian et al |
Keratinogenic differentiation of human ADSCs, modeling the nucleus |
158
|
| 2014 |
Lee et al |
Myogenic differentiation of human mesenchymal stem cells (MSCs) using UV curable poly(urethane acrylate) |
205
|
| 2016 |
Bonakdar et al |
Tenogenic differentiation of stem cells, redifferentiation, and trans-differentiation of tenocytes |
194
|
| 2017 |
Wang et al |
Replication of cancer cells for cell recognition |
209
|
| 2018 |
Kamgouyan et al |
Osteogenic differentiation of human ADSCs, coating of hydroxyapatite on imprinted surface |
200
|
| 2018 |
Farvadi et al |
Nanoparticle uptake on fibroblast imprinted substrate |
210
|
| 2019 |
Kavand et al |
Conductive cell imprinted substrate based on polydimethylsiloxane (PDMS) and carbon nanotube |
211
|
| 2019 |
Kavand et al |
Using chondrocyte as molding template for direct cell photolithography |
165
|
| 2019 |
Moosazadeh et al |
Schwann cell differentiation of rat ADSCs |
206
|
| 2020 |
Gholami et al |
Osteogenic differentiation of MSCs on osteoblast imprinted substrate and regeneration of calvarial bone in rats using collagen-based scaffold |
212
|
| 2020 |
Haramshahi et al |
Tenogenic differentiation induction of the tenocyte imprinted substrate was greater than the tissue replica |
213
|
| 2020 |
Ghazali et al |
Neuronal differentiation of a human stem cell line using ReNcell® |
204
|
| 2021 |
Kashani et al |
Chondrogenic differentiation on a chip and regeneration of cartilage in a rabbit model of hyaline cartilage |
214
|
| 2021 |
Kamgouyan et al |
Determination of nanoscale fingerprints of imprinted substrates |
201
|
| 2021 |
Kashani et al |
Computational fluid dynamics (CFD) simulations on HUVEC, L929 and chondrocyte-imprinted-based integrated microfluidic device |
215
|
| 2021 |
Dadashkhan et al |
Induction of Schwann cell differentiation in human MSCs cultured on imprinted substrates and exposed to β-carotene |
216
|
| 2022 |
Nazbar et al |
Improving stemness in ADSCs cultured on imprinted substrates |
217
|
| 2022 |
Babaei et al. |
Improving cell adhesion, proliferation and osteogenic differentiation in imprinted substrates coated with bone-extracted proteins |
164
|
Bonakdar et al assessed the differentiation, redifferentiation, and transdifferentiation of chondrocyte imprinted substrates. They used the cell imprinted approach to probe stem cell differentiation and sought to determine the commitment of stem cell differentiation to the desired phenotypes. In order to display the potential of cell imprinting to recapitulate the ECM architecture, researchers investigated chondrogenic and tenogenic differentiation and teno-chondro transdifferentiation on a PDMS template of corresponding cells. The results showed that with this simple imprinting procedure, both the nanoscale and microscale aspects of the cell topography and their main phenotypes were transferred to the substrate in a way that the spindle morphologies of semi-fibroblasts and adipose-derived stem cells (ADSCs) were changed into a spherical morphology when cultured on chondrocyte imprinted substrates. The spindle morphology of tenocytes transformed into a spherical morphology when the tenocytes were cultured on chondrocyte imprinted substrates. Based on the results of gene expression analysis, particular genes can be up-regulated when cells are cultured on imprinted substrates. The significant increase in collagen type II expression (as a specific marker of chondrogenic differentiation) in both ADSCs and semi-fibroblast cells indicated the success of the imprinting approach. Similar results were obtained from tenogenic differentiation of a tenocyte imprinted substrate in another study and specific genes for tenocytes such as tenascin, decorin and tenomodulin were adjusted for ADSCs cultured on a tenocyte imprinted substrate.203 In another research, stem cell imprinted substrates were utilized in the long-term expansion of ADSCs to maintain stemness. In addition, downregulation in the expression of specific markers in osteogenic and adipogenic differentiation was observed for stem cells seeded on imprinted substrates that were exposed to the differentiation chemical signals.217
The cell imprinted method provides a monolayer structure that enables cells to develop while maintaining their primary phenotype (Fig. 3). Furthermore, the utilization of growth factors such as TGF-β, FGF, BMP, and IGF, which are commonly suggested for chondrogenesis, is reduced by employing this approach.218 On the other hand, it has been shown that high doses of chemical stimulants may have an inhibitory instead of stimulatory effect. For instance, synovial fibrosis, osteophyte formation, cartilage degeneration and bone remodeling have been reported for altered signaling in TGF-β content.219 Growth factors can assist in expanding proliferation and postponing chondrocyte dedifferentiation. However, their effective concentration in cultures is consistently altered, that means they do not completely prevent phenotypic changes.220
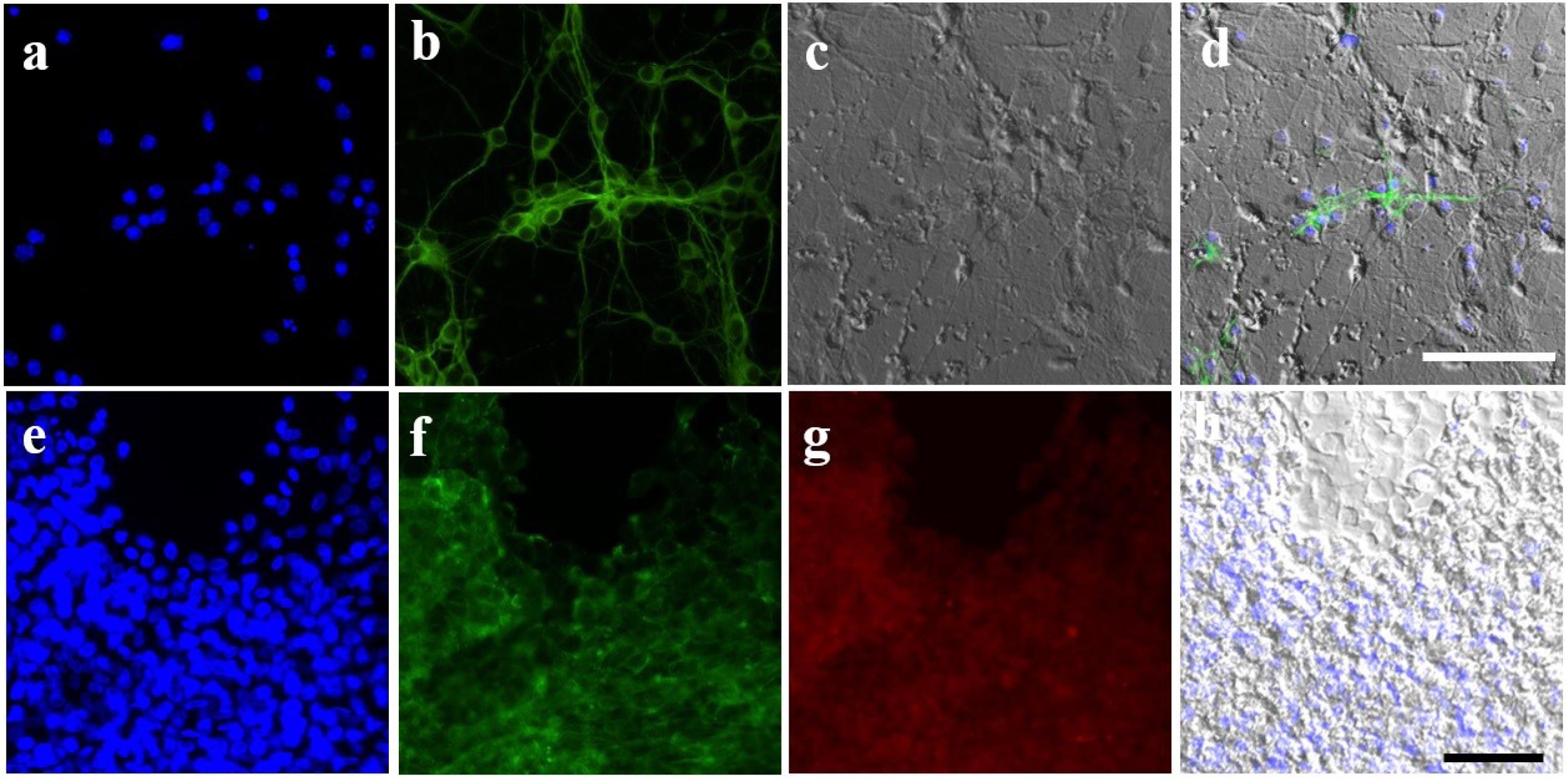
Fig. 3.
Stem cell differentiation on imprinted substrates. a-d) Mouse adipose-derived stem cells (ADSCs) cultured on neuroblast imprinted substrates. a) Nucleus stained with Hoechst, b) actin stained with phalloidin, c) optical image of stem cells and imprinted shapes, and d) merged images. e-h) Human ADSCs cultured on chondrocyte imprinted substrates. e) Nucleus stained with Hoechst, f) actin stained with phalloidin, g) immunostaining of collagen type II, and h) optical image of imprinted shapes.
.
Stem cell differentiation on imprinted substrates. a-d) Mouse adipose-derived stem cells (ADSCs) cultured on neuroblast imprinted substrates. a) Nucleus stained with Hoechst, b) actin stained with phalloidin, c) optical image of stem cells and imprinted shapes, and d) merged images. e-h) Human ADSCs cultured on chondrocyte imprinted substrates. e) Nucleus stained with Hoechst, f) actin stained with phalloidin, g) immunostaining of collagen type II, and h) optical image of imprinted shapes.
Haramshahi et al. reported that tenogenic differentiation potency (scleraxis and tenomodulin expressions) of tenocyte imprinted substrates was higher than the imprinted tendon tissue and lower than BMP12 treated groups.213 Therefore, it is valuable to know the synergistic effect of growth factors (or chemical substances) and cell-imprinted substrates on cell/stem cell functions. It can be predicted that cell imprinted substrates may result in the reduction in the effective concentration of growth factors or decrease the exposure times.
Babaei et al, showed improvement in cell adhesion, proliferation and osteogenic differentiation by coating the osteoblast imprinted substrates with bone extracted proteins.164 In another recently published report, it was found that small molecules such as β-carotene can enhance the differentiation capacity of cell imprinting.216
Overall, the mechanisms of stem cell differentiation induced by physical stimuli and the maintenance of cell phenotype are not realized, though a number of key mechanotransduction components including focal adhesions, cytoskeletal contractility, Rho GTPase signaling and nuclear regulation have been demonstrated to be involved in the force-mediated differentiation.221,222 The cytoskeleton of cells is a dynamic structure, meaning that cells can actively respond to physical signals by adjusting their mechanical properties through the remodeling of cytoskeletal components. This dynamic response of the cytoskeleton not only affects cellular contractility but also triggers molecular signaling pathways, specifically the activation of RhoGTPases. Consequently, these pathways regulate intracellular processes and control gene expression.223
Cell modeling of diseases in vitro
Various models, both continuum and discrete, is granted to depict the aggregate cell behaviors in connection with challenges relating to disease modeling. Normally, discrete cell modeling employs quasi-infinite or boundary less 2D lattices to model collective cell behaviors in Petri dish-like environments. Modeling the entire behavior of cells within in vitro culture environments is a complex subject that is generally carried out by separate cell models, typically cellular automata. Recently, in a particular area of modeling cell behavior, cellular automata have been used for modeling cell adhesion and proliferation.224
An ideal model should imitate all-natural cues in in-vitro condition, including autocrine, paracrine and endocrines as well as physical internal or external signals (e.g., forces, patterns, or electromagnetic waves), which seems impossible to simultaneously consider thousands of factors. In addition to these complexities, genetic diversity, age or nutrition should be added.225 Therefore, simplifying models alongside eliminating factors to the utmost possible extent is rational. Because it has been proven that cell imprinted substrates could mimic the natural topography of cells in vitro, these substrates can be appropriate choices. For example, during the disease progress, both the cell phenotype and expression undergo changes. These cells can be isolated and cultured in vitro to impart imprinted substrates that have exact patterns. Subsequently, adult or embryonic stem cells can be cultured on these imprinted substrates to determine the changes in protein and gene expressions when the cells are exposed to varying concentrations of different drugs and different time points (which is difficult and unethical in animal models). The most important advantages of cell imprinted substrates in disease modeling include the stability of the model and accessibility to the development of the disease during a short period.114,226,227
Various models have been developed to assess diseases in vitro. The appearance of lab- and organ-on-a-chip tools confirms that the information obtained from 2D cell cultures, in Petri dishes, varies significantly from the outcomes obtained from more biomimetic microfluidic environments that were created from interconnected chambers and channels. Therefore, organs-on-chips have a tremendous potential to analyze basic processes of organ physiology and disease. They are well-adapted to the investigation of biological events that rely on tissue microarchitecture and perfusion that include comparatively acute cases of less than one month pathophysiological processes.81 For instance, dynamic alterations of oxygen tension have been applied in organs-on-chips to mimic disease states, including heart ischemia228 or vaso-occlusion in sickle-cell disease as a result of polymerization of hemoglobin S in deoxygenated erythrocytes.229 Based on similar studies, the use of low levels of fluid shear stress similar to those within the collecting ducts and proximal tubules of the living kidney caused elevated differentiation (e.g., epithelial cell polarization, formation of primary cilia), developed molecular and drug delivery roles, and created more in vivo-like toxicity reactions when primary rat,230 dog231 and human232 cells obtained from these tissues were cultured in chips.
Human diseases can easily be investigated in a widely governable environment by using in vitro disease models after the production of characterized human embryonic stem cell lines. The proposing of human‐induced PSCs (iPSCs) a decade ago, paved the way for in vitro research on different diseases.233 The full pluripotency of iPSCs has been explained by various researches via the most accurate experiment of pluripotency (i.e. tetraploid complementation), and revealed the feasibility of obtaining pluripotent iPSCs from somatic cells.234 Due to these properties, iPSCs have multiple biomedical benefits to fundamental studies, drug screening, toxicological studies, disease modeling and cell therapy.235 There has been tremendous progress in disease modeling with the increase of iPSC-derived organoids.236 The soluble and biophysical signs utilized to instruct organoid differentiation from iPSCs have been increasingly purified to produce complex ‘tissues in a dish.237 Besides, researchers have been able to produce specific tissues from organs such as the brain and gastrointestinal tract.238,239
In addition, microvascular network remodeling includes angiogenesis, determined as the germinating of new capillaries and network modeling related to the formation and linkage of available vessels. For instance, blocking remodeling would be useful in numerous pathologies, including cancer, proliferative retinopathies and rheumatoid arthritis. In other diseases such as myocardial infarction, stroke and high blood pressure, strengthening a novel procedure is quite desirable. Improvement of such therapies converges a further comprehension of each sub-process association with microvascular remodeling besides our understanding of how each sub-process is coordinated across a network.240
Alternative methods provide an alternative to conventional strategies and animal models, and they could solve many issues of basic studies and drug development. Since cell imprinted substrates can imitate the natural cell phenotype, researchers can observe different cell viability/toxicity when exposed to substances and cultured on their respective imprints as compared to polystyrene plates.210 The applications of the cell imprinting method are shown schematically in Fig. 4.
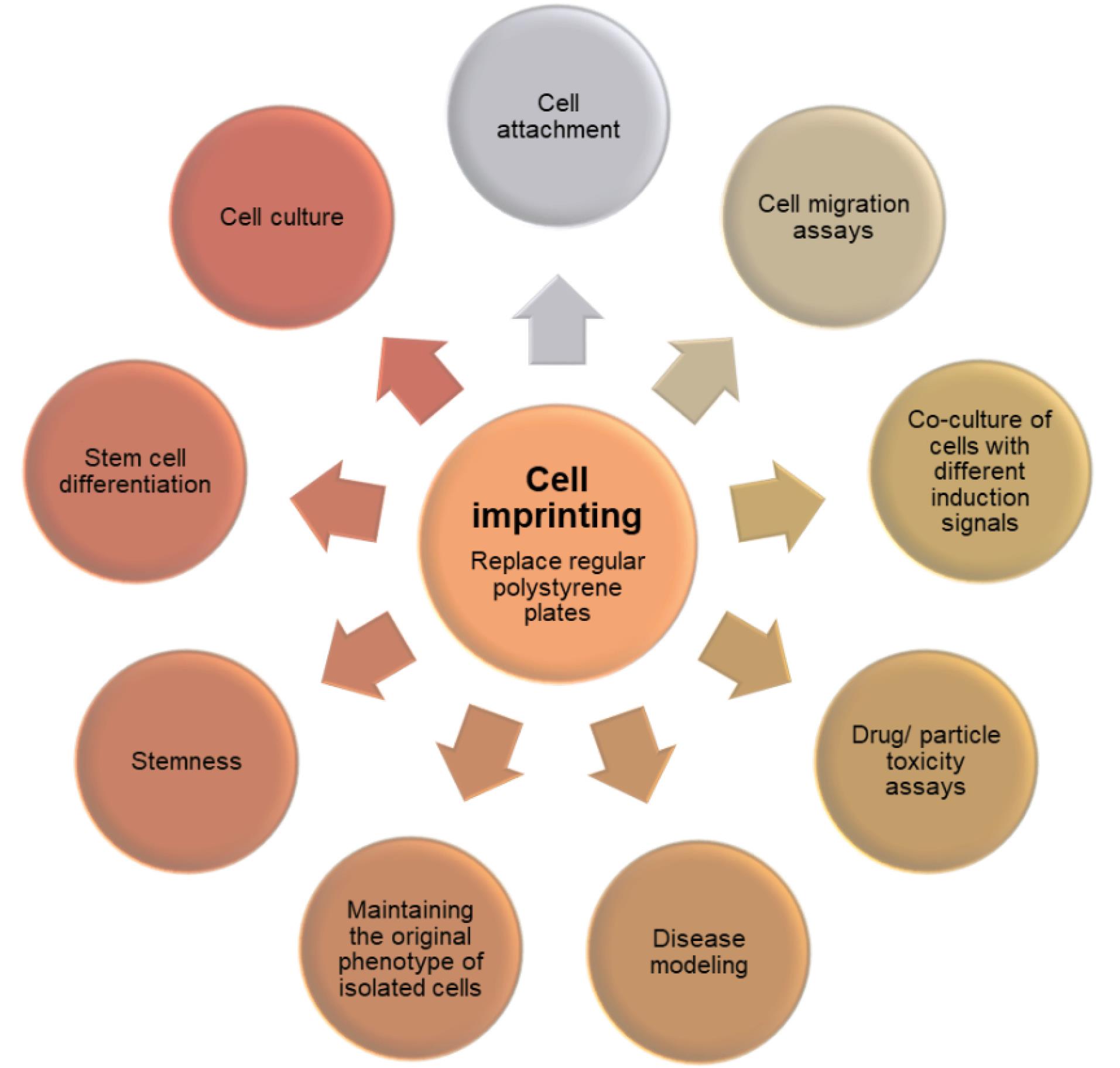
Fig. 4.
The different applications of cell imprinting approach
.
The different applications of cell imprinting approach
Cell therapy
Cell therapy is an approach where cellular components are injected or transplanted into a patient. Throughout the world, many products are presently under extensive analysis and their market is anticipated to grow quickly in the near future.241 Some of the drugs are still present in the form of organic or protein substances to relieve symptoms, not for definitive remedy. However, cell transplantation procedures can be considered to be the frontiers of new technology for fighting against diseases. Cell therapy products utilize cells as a drug. The cells are often obtained from a patient’s own tissues in order to obtain individual attributes by manipulation and culture, and are subsequently re-injected into the patient. Hence, cell therapy products are served in personalized medicine as compared to drugs that can be administered to everyone.242 Cell therapy approaches can be closely linked to gene therapy, cancer vaccines, drug delivery, tissue engineering and regenerative medicine.243 The very basis of these therapies is bone marrow transplantation in cancer treatment.244 They can potentially be used for cardiac diseases (myocardial infarction and heart failure),245 diabetes mellitus,246 bone and joint disorders,247 genetic disorders248 and skin wounds.249 Although each disease has intricate specific features, the basic principles of cell therapy in these cases are almost the same.250
Cell therapy is aimed to repair malfunctions rather than to generate a new tissue, which could bring along duplicity and unfavorable consequences. Different types of cells can be applied to heal the defective tissue such as resident stem cells, multipotent adult progenitor cells or embryonic stem cells. There are various procedures that include the use of both autologous (from the same individual) and allogeneic (from another individual of the same species) progenitors of adult or embryonic stem cells and different trophic factors such as VEGF, TGF,EPO or HGF, which aim to stimulate proliferation and differentiation of resident stem cells.250
Despite its vast benefits, problems exist with cell therapy that should be solved. The most critical limitations of stem cell therapy are safety issues, including tumorigenicity and immunogenicity. Although the risks are considerably lower in cell (non-stem cell) therapy, the improvement in the quality of a patient’s life after cell transplantation should be guaranteed.251
Cell imprinted substrates can induce physical signals for the redifferentiation of phenotypically altered cells in vitro. Considering the increased safety of the induction by physical cues rather than chemical cues, cell imprinting can be used for many kinds of cell therapeutic applications in which the source of normal juvenile cells is not available in an older patient. Because the potency of normal cells is reduced in an older person, it might be desirable to isolate cells from an older patient and culture them on imprinted substrates derived from young brain-dead donors or fetal-derived cells. In addition, for therapeutic procedures such as autologous chondrocyte transplantation that require millions of cells through expansions, cell-imprinted substrates could be a desirable substitute for conventional tissue culture polystyrene plates. However, the proposed concepts should be researched in the future. Recently, Kashani et al fabricated a chip with cell-imprinted topography at the bottom to culture stem cells under a dynamic culture for 14 days. The differentiated stem cells by the chondrocyte imprinted substrate could regenerate hyaline cartilage in a rabbit model when compared to undifferentiated stem cells.214 This finding was important since it was previously reported that undifferentiated stem cells could not regenerate meniscus tissue after transplantation.252 In other words, in order to regenerate tissues, pre-stimulation of stem cells is required prior to transplantation.
Conclusion and future perspectives
Cell imprinted substrates could be used to investigate the molecular processes of organ growth and diseases by novel methods in vitro apart from the interplay of the body with drugs, environmental agents, consumer products and medical tools. The idea of modeling disease by cell imprinting should be analyzed and compared with studies on animal models in the future. In addition, it is suggested that both physical and chemical signals should be combined for better simulation of the natural environment. Surface coating of the cell imprinted substrate with ECM molecules such as collagen or the addition of small molecules to the medium used to culture the cells could be considered as a subject of future research. As illustrated in the current study, microfluidic chips with cell-imprinted patterns can be fabricated and utilized for dynamic cultures. Improvements in cell imprinting techniques can pave the way for pre-differentiation of stem cells towards the desired phenotype through a safe, efficient, repeatable and reliable approach.
Review Highlights
What is the current knowledge?
√ For cell-based experiments involved in research or clinics, an artificial ECM is required to support cell function, which can be considered as a substrate.
√ To better mimic the natural environment in vitro, factors such as forced polarity, flattened cell shape, mechanical/biochemical signals, and subsequent cell-to-cell communication must be regulated.
What is new here?
√ The various 3D cell culture techniques such as organoids, pellets, spheroids, 3D scaffolds and hydrogels can be used as simulate in vivo conditions for cell growth.
√ The cells show a greater inclination to attach to rough surfaces rather than smooth ones.
√ It is apparent that changes in the shape of a nucleus will change the cell’s fate.
√ Cell imprinted substrates can induce physical signals for the redifferentiation of phenotypically altered cells in vitro.
√ Imprinted polymers are considered to be artificial receptors or plastic antibodies, which can mimic the biological role of a natural receptor or antibody.
Acknowledgement
We would like to thank Pasteur Institute and Motamed Cancer Institute of Iran for supporting this research. We would also like to thank our colleagues at Royan Institute and University of Aveiro who gave insight and knowledge that considerably aided the research.
Competing Interests
The authors declare no conflict interests.
Ethical Statement
Not applicable.
Funding
The authors have no funding to disclose.
References
- Barthes J, Özçelik H, Hindié M, Ndreu-Halili A, Hasan A, Vrana NE. Cell microenvironment engineering and monitoring for tissue engineering and regenerative medicine: the recent advances. Biomed Res Int 2014; 2014. 10.1155/2014/921905.
- Tabata Y. Biomaterials design of culture substrates for cell research. Inflamm Regen 2011; 31:137-45. doi: 10.2492/inflammregen.31.137 [Crossref] [ Google Scholar]
- Heydari T, Heidari M, Mashinchian O, Wojcik M, Xu K, Dalby MJ. Development of a virtual cell model to predict cell response to substrate topography. ACS Nano 2017; 11:9084-92. doi: 10.1021/acsnano.7b03732 [Crossref] [ Google Scholar]
- Kashani SY, Moraveji MK, Bonakdar S. Computational and experimental studies of a cell-imprinted-based integrated microfluidic device for biomedical applications. Sci Rep 2021; 11:1-17. doi: 10.1038/s41598-021-91616-2 [Crossref] [ Google Scholar]
- Ferrari M, Cirisano F, Morán MC. Mammalian cell behavior on hydrophobic substrates: influence of surface properties. Colloids Interface 2019; 3:48. doi: 10.3390/colloids3020048 [Crossref] [ Google Scholar]
- Xing F, Li L, Zhou C, Long C, Wu L, Lei H, et al. Regulation and Directing Stem Cell Fate by Tissue Engineering Functional Microenvironments: Scaffold Physical and Chemical Cues. Stem Cells Int 2019; 2019. 10.1155/2019/2180925.
- Kapałczyńska M, Kolenda T, Przybyła W, Zajączkowska M, Teresiak A, Filas V. 2D and 3D cell cultures - a comparison of different types of cancer cell cultures. Arch Med Sci 2018; 14:910-9. doi: 10.5114/aoms.2016.63743 [Crossref] [ Google Scholar]
- Habanjar O, Diab-Assaf M, Caldefie-Chezet F, Delort L. 3D Cell Culture Systems: Tumor Application, Advantages, and Disadvantages. Int J Mol Sci 2021; 22:1-35. doi: 10.3390/ijms222212200 [Crossref] [ Google Scholar]
- Ajjarapu SM, Tiwari A, Kumar S. Applications and Utility of Three-Dimensional In Vitro Cell Culture for Therapeutics. Future Pharm 2023; 3:213-28. doi: 10.3390/futurepharmacol3010015 [Crossref] [ Google Scholar]
- Stratton S, Shelke NB, Hoshino K, Rudraiah S, Kumbar SG. Bioactive polymeric scaffolds for tissue engineering. Bioact Mater 2016; 1:93-108. doi: 10.1016/j.bioactmat.2016.11.001 [Crossref] [ Google Scholar]
- Shahriyari F, Janmaleki M, Sharifi S, Hesar ME, Hoshian S, Taghiabadi R, et al. Effect of cell imprinting on viability and drug susceptibility of breast cancer cells to doxorubicin. Acta Biomater 2020. 113: 119-29. 10.1016/j.actbio.2020.06.007.
- Anaya J-M, Shoenfeld Y, Rojas-Villarraga A, Levy RA, Cervera R. Autoimmunity: from bench to bedside. El Rosario University Press; 2013.
- Yao T, Asayama Y. Animal‐cell culture media: History, characteristics, and current issues. Reprod Med Biol 2017; 16:99-117. doi: 10.1002/rmb2.12024 [Crossref] [ Google Scholar]
- Ryan J. Introduction to animal cell culture: Technical bulletin. Lowell, MA: Corning Incorporated Life Sci; 2008.
- Sanyal S. Culture and assay systems used for 3D cell culture. Corning 2014; 9:1-18. [ Google Scholar]
- Kapałczyńska M, Kolenda T, Przybyła W, Zajączkowska M, Teresiak A, Filas V. 2D and 3D cell cultures–a comparison of different types of cancer cell cultures. Arch Med Sci 2018; 14:910. doi: 10.5114/aoms.2016.63743 [Crossref] [ Google Scholar]
- Breslin S, O’Driscoll L. Three-dimensional cell culture: the missing link in drug discovery. Drug Discov Today 2013; 18:240-9. doi: 10.1016/j.drudis.2012.10.003 [Crossref] [ Google Scholar]
- Alves NM, Pashkuleva I, Reis RL, Mano JF. Controlling cell behavior through the design of polymer surfaces. Small 2010; 6:2208-20. doi: 10.1002/smll.201000233 [Crossref] [ Google Scholar]
- Hoarau-Véchot J, Rafii A, Touboul C, Pasquier J. Halfway between 2D and animal models: are 3D cultures the ideal tool to study cancer-microenvironment interactions?. Int J Mol Sci 2018; 19:181. doi: 10.3390/ijms19010181 [Crossref] [ Google Scholar]
- Yamada KM, Cukierman E. Modeling tissue morphogenesis and cancer in 3D. Cell 2007; 130:601-10. doi: 10.1016/j.cell.2007.08.006 [Crossref] [ Google Scholar]
- Duval K, Grover H, Han L-H, Mou Y, Pegoraro AF, Fredberg J. Modeling physiological events in 2D vs3D cell culture. Physiology 2017; 32:266-77. doi: 10.1152/physiol.00036.2016 [Crossref] [ Google Scholar]
- Baker BM, Chen CS. Deconstructing the third dimension–how 3D culture microenvironments alter cellular cues. J Cell Sci 2012; 125:3015-24. doi: 10.1242/jcs.079509 [Crossref] [ Google Scholar]
- Bissell MJ, Rizki A, Mian IS. Tissue architecture: the ultimate regulator of breast epithelial function. Curr Opin Cell Biol 2003; 15:753. doi: 10.1016/j.ceb.2003.10.016 [Crossref] [ Google Scholar]
- Hickman JA, Graeser R, de Hoogt R, Vidic S, Brito C, Gutekunst M. Three‐dimensional models of cancer for pharmacology and cancer cell biology: capturing tumor complexity in vitro/ex vivo. Biotechnol J 2014; 9:1115-28. doi: 10.1002/biot.201300492 [Crossref] [ Google Scholar]
- Mahmud G, Campbell CJ, Bishop KJ, Komarova YA, Chaga O, Soh S. Directing cell motions on micropatterned ratchets. Nat Phys 2009; 5:606-12. doi: 10.1038/nphys1306 [Crossref] [ Google Scholar]
- Kilian KA, Bugarija B, Lahn BT, Mrksich M. Geometric cues for directing the differentiation of mesenchymal stem cells. PNAS 2010; 107:4872-7. doi: 10.1073/pnas.0903269107 [Crossref] [ Google Scholar]
- Nelson CM, Bissell MJ. Of extracellular matrix, scaffolds, and signaling: tissue architecture regulates development, homeostasis, and cancer. Annu Rev Cell Dev Biol 2006; 22:287-309. doi: 10.1146/annurev.cellbio.22.010305.104315 [Crossref] [ Google Scholar]
- Meyers J, Craig J, Odde DJ. Potential for control of signaling pathways via cell size and shape. Curr Biol 2006; 16:1685-93. doi: 10.1016/j.cub.2006.07.056 [Crossref] [ Google Scholar]
- Pampaloni F, Reynaud EG, Stelzer EH. The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol 2007; 8:839-45. doi: 10.1038/nrm2236 [Crossref] [ Google Scholar]
- Gilbert PM, Havenstrite KL, Magnusson KE, Sacco A, Leonardi NA, Kraft P. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science 2010; 329:1078-81. doi: 10.1126/science.1191035 [Crossref] [ Google Scholar]
- Fischbach C, Chen R, Matsumoto T, Schmelzle T, Brugge JS, Polverini PJ. Engineering tumors with 3D scaffolds. Nat Methods 2007; 4:855-60. doi: 10.1038/nmeth1085 [Crossref] [ Google Scholar]
- Monteiro MV, Gaspar VM, Ferreira LP, Mano JF. Hydrogel 3D in vitro tumor models for screening cell aggregation mediated drug response. Biomate Sci 2020; 8:1855-64. doi: 10.1039/c9bm02075f [Crossref] [ Google Scholar]
- Abdollahi S. Extracellular vesicles from organoids and 3d culture systems. Biotechnol Bioeng 2021; 118:1029-49. doi: 10.1002/bit.27606 [Crossref] [ Google Scholar]
- Simian M, Bissell MJ. Organoids: a historical perspective of thinking in three dimensions. J Cell Biol 2017; 216:31-40. doi: 10.1083/jcb.201610056 [Crossref] [ Google Scholar]
- Huch M, Koo B-K. Modeling mouse and human development using organoid cultures. Development 2015. 142: 3113-25. 10.1242/dev.118570.
- Clevers H. Modeling development and disease with organoids. Cell 2016; 165:1586-97. doi: 10.1016/j.cell.2016.05.082 [Crossref] [ Google Scholar]
- Dutta D, Heo I, Clevers H. Disease modeling in stem cell-derived 3D organoid systems. Trends Mol Med 2017; 23:393-410. doi: 10.1016/j.molmed.2017.02.007 [Crossref] [ Google Scholar]
- Es HA, Montazeri L, Aref AR, Vosough M, Baharvand H. Personalized cancer medicine: an organoid approach. Trends Biotechnol 2018; 36:358-71. doi: 10.1016/j.tibtech.2017.12.005 [Crossref] [ Google Scholar]
- Zanoni M, Cortesi M, Zamagni A, Arienti C, Pignatta S, Tesei A. Modeling neoplastic disease with spheroids and organoids. J Hematol Oncol 2020; 13:1-15. doi: 10.1186/s13045-020-00931-0 [Crossref] [ Google Scholar]
- Xu H, Lyu X, Yi M, Zhao W, Song Y, Wu K. Organoid technology and applications in cancer research. J hematol oncol 2018; 11:1-15. doi: 10.1186/s13045-018-0662-9 [Crossref] [ Google Scholar]
- Yoon WH, Lee H-R, Kim S, Kim E, Ku JH, Shin K. Use of inkjet-printed single cells to quantify intratumoral heterogeneity. Biofabrication 2020; 12:035030. doi: 10.1088/1758-5090/ab9491 [Crossref] [ Google Scholar]
- Tatullo M, Marrelli B, Benincasa C, Aiello E, Makeeva I, Zavan B. Organoids in translational oncology. J Clin Med 2020; 9:2774. doi: 10.3390/jcm9092774 [Crossref] [ Google Scholar]
- Xu H, Jiao D, Liu A, Wu K. Tumor organoids: applications in cancer modeling and potentials in precision medicine. J Hematol Oncol 2022; 15:58. doi: 10.1186/s13045-022-01278-4 [Crossref] [ Google Scholar]
- Lv D, Hu Z, Lu L, Lu H, Xu X. Three-dimensional cell culture: A powerful tool in tumor research and drug discovery. Oncol Let 2017; 14:6999-7010. doi: 10.3892/ol.2017.7134 [Crossref] [ Google Scholar]
- Narmi MT, Shoja HM, Haiaty S, Mahdipour M, Rahbarghazi R. Melatonin blunted the angiogenic activity in 3D colon cancer tumoroids by the reduction of endocan. Cancer Cell Int 2023; 23:118. doi: 10.1186/s12935-023-02951-5 [Crossref] [ Google Scholar]
- Mead BE, Karp JM. All models are wrong, but some organoids may be useful. Genome Biol 2019; 20:66. doi: 10.1186/s13059-019-1677-4 [Crossref] [ Google Scholar]
- Huch M, Gehart H, Van Boxtel R, Hamer K, Blokzijl F, Verstegen MM. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 2015; 160:299-312. doi: 10.1016/j.cell.2014.11.050 [Crossref] [ Google Scholar]
- Shen H. Core Concept: Organoids have opened avenues into investigating numerous diseases. But how well do they mimic the real thing? PNAS 2018; 115:3507-9. doi: 10.1073/pnas.1803647115 [Crossref] [ Google Scholar]
- Watson CL, Mahe MM, Múnera J, Howell JC, Sundaram N, Poling HM. An in vivo model of human small intestine using pluripotent stem cells. Nat Med 2014; 20:1310. doi: 10.1038/nm.3737 [Crossref] [ Google Scholar]
- Caron MM, Emans PJ, Coolsen MM, Voss L, Surtel DA, Cremers A. Redifferentiation of dedifferentiated human articular chondrocytes: comparison of 2D and 3D cultures. Osteoarthr Cartil 2012; 20:1170-8. doi: 10.1016/j.joca.2012.06.016 [Crossref] [ Google Scholar]
- Grigull NP, Redeker JI, Schmitt B, Saller MM, Schönitzer V, Mayer-Wagner S. Chondrogenic Potential of Pellet Culture Compared to High-Density Culture on a Bacterial Cellulose Hydrogel. Int J Mol Sci 2020; 21:2785. doi: 10.3390/ijms21082785 [Crossref] [ Google Scholar]
- Gilvari H, de Jong W, Schott DL. Quality parameters relevant for densification of bio-materials: Measuring methods and affecting factors-A review. Biomass and Bioenergy 2019; 120:117-34. doi: 10.1016/j.biombioe.2018.11.013 [Crossref] [ Google Scholar]
- Zhang L, Su P, Xu C, Yang J, Yu W, Huang D. Chondrogenic differentiation of human mesenchymal stem cells: a comparison between micromass and pellet culture systems. Biotechnol Let 2010; 32:1339-46. doi: 10.1007/s10529-010-0293-x [Crossref] [ Google Scholar]
- Edmondson R, Broglie JJ, Adcock AF, Yang L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev Technol 2014; 12:207-18. doi: 10.1089/adt.2014.573 [Crossref] [ Google Scholar]
- Oliveira MB, Neto AI, Correia CR, Rial-Hermida MI, Alvarez-Lorenzo C, Mano JF. Superhydrophobic chips for cell spheroids high-throughput generation and drug screening. ACS appl Mater Interfaces 2014; 6:9488-95. doi: 10.1021/am5018607 [Crossref] [ Google Scholar]
- Liu D, Chen S, Win Naing M. A review of manufacturing capabilities of cell spheroid generation technologies and future development. Biotechnol Bioeng 2021; 118:542-54. doi: 10.1002/bit.27620 [Crossref] [ Google Scholar]
- Langhans SA. Three-dimensional in vitro cell culture models in drug discovery and drug repositioning. Front Pharmacol 2018; 9:6. doi: 10.3389/fphar.2018.00006 [Crossref] [ Google Scholar]
- Torisawa Y-s, Takagi A, Shiku H, Yasukawa T, Matsue T. A multicellular spheroid-based drug sensitivity test by scanning electrochemical microscopy. Oncol Rep 2005; 13:1107-12. [ Google Scholar]
- Mehta G, Hsiao AY, Ingram M, Luker GD, Takayama S. Opportunities and challenges for use of tumor spheroids as models to test drug delivery and efficacy. JCR 2012; 164:192-204. doi: 10.1016/j.jconrel.2012.04.045 [Crossref] [ Google Scholar]
- Ryu N-E, Lee S-H, Park H. Spheroid Culture System Methods and Applications for Mesenchymal Stem Cells. Cells 2019; 8:1620. doi: 10.3390/cells8121620 [Crossref] [ Google Scholar]
- Cui X, Hartanto Y, Zhang H. Advances in multicellular spheroids formation. J R Soc Interface 2017; 14:20160877. doi: 10.1098/rsif.2016.0877 [Crossref] [ Google Scholar]
- Neto A, Correia C, Oliveira M, Rial-Hermida M, Alvarez-Lorenzo C, Reis R. A novel hanging spherical drop system for the generation of cellular spheroids and high throughput combinatorial drug screening. Biomaterials Science 2015; 3:581-5. doi: 10.1039/c4bm00411f [Crossref] [ Google Scholar]
- DJ LKM. Hydrogels for tissue engeneering. Chem Rev 2001; 101:1869-80. doi: 10.1021/cr000108x [Crossref] [ Google Scholar]
- Bryant SJ, Anseth KS. The effects of scaffold thickness on tissue engineered cartilage in photocrosslinked poly (ethylene oxide) hydrogels. Biomaterials 2001; 22:619-26. doi: 10.1016/s0142-9612(00)00225-8 [Crossref] [ Google Scholar]
- Tabata Y. Tissue regeneration based on growth factor release. Tissue Eng 2003; 9:5-15. doi: 10.1089/10763270360696941 [Crossref] [ Google Scholar]
- El-Sherbiny IM, Yacoub MH. Hydrogel scaffolds for tissue engineering: Progress and challenges. Glob Cardiol Sci Pract 2013; 2013:38. doi: 10.5339/gcsp.2013.38 [Crossref] [ Google Scholar]
- Ju HK, Kim SY, Kim SJ, Lee YM. pH/temperature‐responsive semi‐IPN hydrogels composed of alginate and poly (N‐isopropylacrylamide). J Appl Polym Sci 2002; 83:1128-39. doi: 10.1002/app.10137 [Crossref] [ Google Scholar]
- Chaudhari AA, Vig K, Baganizi DR, Sahu R, Dixit S, Dennis V. Future prospects for scaffolding methods and biomaterials in skin tissue engineering: a review. Int J Mol Sci 2016; 17:1974. doi: 10.3390/ijms17121974 [Crossref] [ Google Scholar]
- Bahram M, Mohseni N, Moghtader M. An introduction to hydrogels and some recent applications. Emerging concepts in analysis and applications of hydrogels. IntechOpen; 2016. 10.5772/64301.
- Costa AM, Mano JF. Extremely strong and tough hydrogels as prospective candidates for tissue repair–A review. Eur Polym J 2015; 72:344-64. doi: 10.1016/j.eurpolymj.2015.07.053 [Crossref] [ Google Scholar]
- Zimmermann H, Shirley SG, Zimmermann U. Alginate-based encapsulation of cells: past, present, and future. Curr Diab Rep 2007; 7:314-20. doi: 10.1007/s11892-007-0051-1 [Crossref] [ Google Scholar]
- Caliari SR, Burdick JA. A practical guide to hydrogels for cell culture. Nat Methods 2016; 13:405-14. doi: 10.1038/nmeth.3839 [Crossref] [ Google Scholar]
- Correia CR, Nadine S, Mano JF. Cell encapsulation systems toward modular tissue regeneration: from immunoisolation to multifunctional devices. Adv Func Mater 2020; 30:1908061. doi: 10.1002/adfm.201908061 [Crossref] [ Google Scholar]
- Correia CR, Reis RL, Mano JoF. Multilayered hierarchical capsules providing cell adhesion sites. Biomacromolecules 2013; 14:743-51. doi: 10.1021/bm301833z [Crossref] [ Google Scholar]
- Correia CR, Ghasemzadeh-Hasankolaei M, Mano JF. Cell encapsulation in liquified compartments: Protocol optimization and challenges. Plos One 2019; 14:e0218045. doi: 10.1371/journal.pone.0218045 [Crossref] [ Google Scholar]
- Shahabipour F, Ashammakhi N, Oskuee RK, Bonakdar S, Hoffman T, Shokrgozar MA. Key components of engineering vascularized 3-dimensional bioprinted bone constructs. Transl Res 2020; 216:57-76. doi: 10.1016/j.trsl.2019.08.010 [Crossref] [ Google Scholar]
- Fazal F, Raghav S, Callanan A, Koutsos V, Radacsi N. Recent advancements in the bioprinting of vascular grafts. Biofabrication 2021. 13: 1-18. 10.1088/1758-5090/ac0963.
- Zhang Z, Nagrath S. Microfluidics and cancer: are we there yet?. Biomed Microdevices 2013; 15:595-609. doi: 10.1007/s10544-012-9734-8 [Crossref] [ Google Scholar]
- Velve-Casquillas G, Le Berre M, Piel M, Tran PT. Microfluidic tools for cell biological research. Nano Today 2010; 5:28-47. doi: 10.1016/j.nantod.2009.12.001 [Crossref] [ Google Scholar]
- Mandrycky C, Phong K, Zheng Y. Tissue engineering toward organ-specific regeneration and disease modeling. MRS Comm 2017; 7:332-47. doi: 10.1557/mrc.2017.58 [Crossref] [ Google Scholar]
- Bhatia SN, Ingber DE. Microfluidic organs-on-chips. Nat Biotechnol 2014; 32:760. doi: 10.1038/nbt.2989 [Crossref] [ Google Scholar]
- Oliveira NM, Reis RL, Mano JF. Open Fluidics: A Cell Culture Flow System Developed Over Wettability Contrast‐Based Chips. Adv Healthc Mater 2017; 6:1700638. doi: 10.1002/adhm.201700638 [Crossref] [ Google Scholar]
- Gale BK, Jafek AR, Lambert CJ, Goenner BL, Moghimifam H, Nze UC. A review of current methods in microfluidic device fabrication and future commercialization prospects. Inventions 2018; 3:60. doi: 10.3390/inventions3030060 [Crossref] [ Google Scholar]
- Sousa MP, Mano JF. Patterned superhydrophobic paper for microfluidic devices obtained by writing and printing. Cellulose 2013; 20:2185-90. doi: 10.1007/s10570-013-9991-6 [Crossref] [ Google Scholar]
- Torino S, Corrado B, Iodice M, Coppola G. Pdms-based microfluidic devices for cell culture. Inventions 2018; 3:65. doi: 10.3390/inventions3030065 [Crossref] [ Google Scholar]
- Gupta N, Liu JR, Patel B, Solomon DE, Vaidya B, Gupta V. Microfluidics‐based 3D cell culture models: Utility in novel drug discovery and delivery research. Bioeng Transl Med 2016; 1:63-81. doi: 10.1002/btm2.10013 [Crossref] [ Google Scholar]
- Damiati S, Kompella UB, Damiati SA, Kodzius R. Microfluidic devices for drug delivery systems and drug screening. Genes 2018; 9:103. doi: 10.3390/genes9020103 [Crossref] [ Google Scholar]
- Beckwitt CH, Clark AM, Wheeler S, Taylor DL, Stolz DB, Griffith L. Liver ‘organ on a chip’. Exp Cell Res 2018; 363:15-25. doi: 10.1016/j.yexcr.2017.12.023 [Crossref] [ Google Scholar]
- Prentice-Mott HV, Chang C-H, Mahadevan L, Mitchison TJ, Irimia D, Shah JV. Biased migration of confined neutrophil-like cells in asymmetric hydraulic environments. PNAS 2013; 110:21006-11. doi: 10.1073/pnas.1317441110 [Crossref] [ Google Scholar]
- Radisic M, Deen W, Langer R, Vunjak-Novakovic G. Mathematical model of oxygen distribution in engineered cardiac tissue with parallel channel array perfused with culture medium containing oxygen carriers. Am J Physiol Heart Circ Physiol 2005; 288:H1278-H89. doi: 10.1152/ajpheart.00787.2004 [Crossref] [ Google Scholar]
- Xiao R-R, Zeng W-J, Li Y-T, Zou W, Wang L, Pei X-F. Simultaneous generation of gradients with gradually changed slope in a microfluidic device for quantifying axon response. Anal Chem 2013; 85:7842-50. doi: 10.1021/ac4022055 [Crossref] [ Google Scholar]
- Peng C-C, Liao W-H, Chen Y-H, Wu C-Y, Tung Y-C. A microfluidic cell culture array with various oxygen tensions. Lab Chip 2013; 13:3239-45. doi: 10.1039/c3lc50388g [Crossref] [ Google Scholar]
- Cimetta E, Cannizzaro C, James R, Biechele T, Moon RT, Elvassore N. Microfluidic device generating stable concentration gradients for long term cell culture: application to Wnt3a regulation of β-catenin signaling. Lab Chip 2010; 10:3277-83. doi: 10.1039/c0lc00033g [Crossref] [ Google Scholar]
- Seidi A, Kaji H, Annabi N, Ostrovidov S, Ramalingam M, Khademhosseini A. A microfluidic-based neurotoxin concentration gradient for the generation of an in vitro model of Parkinson’s disease. Biomicrofluidics 2011; 5:022214. doi: 10.1063/1.3580756 [Crossref] [ Google Scholar]
- Takayama S, Ostuni E, LeDuc P, Naruse K, Ingber DE, Whitesides GM. Subcellular positioning of small molecules. Nature 2001; 411:1016. doi: 10.1038/35082637 [Crossref] [ Google Scholar]
- Chen S, Lee LP. Non-invasive microfluidic gap junction assay. Integr Biol 2010; 2:130-8. doi: 10.1039/b919392h [Crossref] [ Google Scholar]
- Viravaidya K, Shuler ML. Incorporation of 3T3‐L1 cells to mimic bioaccumulation in a microscale cell culture analog device for toxicity studies. Biotechnol Prog 2004; 20:590-7. doi: 10.1021/bp034238d [Crossref] [ Google Scholar]
- Sin A, Chin KC, Jamil MF, Kostov Y, Rao G, Shuler ML. The design and fabrication of three‐chamber microscale cell culture analog devices with integrated dissolved oxygen sensors. Biotechnol Prog 2004; 20:338-45. doi: 10.1021/bp034077d [Crossref] [ Google Scholar]
- Sung JH, Kam C, Shuler ML. A microfluidic device for a pharmacokinetic–pharmacodynamic (PK–PD) model on a chip. Lab Chip 2010; 10:446-55. doi: 10.1039/b917763a [Crossref] [ Google Scholar]
- Zheng W, Wang Z, Zhang W, Jiang X. A simple PDMS-based microfluidic channel design that removes bubbles for long-term on-chip culture of mammalian cells. Lab Chip 2010; 10:2906-10. doi: 10.1039/c005274d [Crossref] [ Google Scholar]
- Domansky K, Leslie DC, McKinney J, Fraser JP, Sliz JD, Hamkins-Indik T. Clear castable polyurethane elastomer for fabrication of microfluidic devices. Lab Chip 2013; 13:3956-64. doi: 10.1039/c3lc50558h [Crossref] [ Google Scholar]
- Montazeri L, Bonakdar S, Taghipour M, Renaud P, Baharvand H. Modification of PDMS to fabricate PLGA microparticles by a double emulsion method in a single microfluidic device. Lab Chip 2016; 16:2596-600. doi: 10.1039/C6LC00437G [Crossref] [ Google Scholar]
- Tehranirokh M, Kouzani AZ, Francis PS, Kanwar JR. Microfluidic devices for cell cultivation and proliferation. Biomicrofluidics 2013; 7:051502. doi: 10.1063/1.4826935 [Crossref] [ Google Scholar]
- Fung Y-c. Biomechanics: mechanical properties of living tissues. Springer Science & Business Media; 2013.
- Chan B, Leong K. Scaffolding in tissue engineering: general approaches and tissue-specific considerations. Eur Spine J 2008; 17:467-79. doi: 10.1007/s00586-008-0745-3 [Crossref] [ Google Scholar]
- Choi JR, Yong KW, Choi JY. Effects of mechanical loading on human mesenchymal stem cells for cartilage tissue engineering. J Cellular Physiol 2018; 233:1913-28. doi: 10.1002/jcp.26018 [Crossref] [ Google Scholar]
- Custódio CA, Reis RL, Mano JF. Engineering biomolecular microenvironments for cell instructive biomaterials. Adv Healthc Mater 2014; 3:797-810. doi: 10.1002/adhm.201300603 [Crossref] [ Google Scholar]
- Alvarez-Lorenzo C, Concheiro A. Smart drug delivery systems: from fundamentals to the clinic. Chem Comm 2014; 50:7743-65. [ Google Scholar]
- Chen L, Wang X, Lu W, Wu X, Li J. Molecular imprinting: perspectives and applications. Chemical Soc Rev 2016; 45:2137-211. doi: 10.1039/C6CS00061D [Crossref] [ Google Scholar]
- Ermis M, Antmen E, Hasirci V. Micro and Nanofabrication methods to control cell-substrate interactions and cell behavior: A review from the tissue engineering perspective. Bioact Mater 2018; 3:355-69. doi: 10.1016/j.bioactmat.2018.05.005 [Crossref] [ Google Scholar]
- Wang S, Li J, Zhou Z, Zhou S, Hu Z. Micro-/nano-scales direct cell behavior on biomaterial surfaces. Molecules 2019. 24: 75. 10.3390/molecules24010075.
- Bjørge IM, Choi IS, Correia CR, Mano JF. Nanogrooved microdiscs for bottom-up modulation of osteogenic differentiation. Nanoscale 2019; 11:16214-21. doi: 10.1039/C9NR06267J [Crossref] [ Google Scholar]
- Gentile F, Tirinato L, Battista E, Causa F, Liberale C, di Fabrizio EM. Cells preferentially grow on rough substrates. Biomaterials 2010; 31:7205-12. doi: 10.1016/j.biomaterials.2010.06.016 [Crossref] [ Google Scholar]
- Mahmoudi M, Bonakdar S, Shokrgozar MA, Aghaverdi H, Hartmann R, Pick A. Cell-imprinted substrates direct the fate of stem cells. ACS Nano 2013; 7:8379-84. doi: 10.1021/nn403844q [Crossref] [ Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell 2006; 126:677-89. doi: 10.1016/j.cell.2006.06.044 [Crossref] [ Google Scholar]
- McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell 2004; 6:483-95. doi: 10.1016/s1534-5807(04)00075-9 [Crossref] [ Google Scholar]
- Biggs MJ, Richards RG, Gadegaard N, Wilkinson CD, Oreffo RO, Dalby MJ. The use of nanoscale topography to modulate the dynamics of adhesion formation in primary osteoblasts and ERK/MAPK signalling in STRO-1+ enriched skeletal stem cells. Biomaterials 2009; 30:5094-103. doi: 10.1016/j.biomaterials.2009.05.049 [Crossref] [ Google Scholar]
- Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol 2006; 7:265-75. doi: 10.1038/nrm1890 [Crossref] [ Google Scholar]
- Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. PNAS 1997; 94:849-54. doi: 10.1073/pnas.94.3.849 [Crossref] [ Google Scholar]
- Ingber DE. Tensegrity: the architectural basis of cellular mechanotransduction. Annu Rev Physiol 1997; 59:575-99. doi: 10.1146/annurev.physiol.59.1.575 [Crossref] [ Google Scholar]
- Fedorchak GR, Kaminski A, Lammerding J. Cellular mechanosensing: getting to the nucleus of it all. Prog Biophys Mol Biol 2014; 115:76-92. doi: 10.1016/j.pbiomolbio.2014.06.009 [Crossref] [ Google Scholar]
- Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat Rev Mol Cell Biol 2009; 10:75-82. doi: 10.1038/nrm2594 [Crossref] [ Google Scholar]
- Guilluy C, Osborne LD, Van Landeghem L, Sharek L, Superfine R, Garcia-Mata R. Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nat Cell Biol 2014; 16:376-81. doi: 10.1038/ncb2927 [Crossref] [ Google Scholar]
- Belaadi N, Aureille J, Guilluy C. Under pressure: mechanical stress management in the nucleus. Cells 2016; 5:27. doi: 10.3390/cells5020027 [Crossref] [ Google Scholar]
- Anselme K, Wakhloo NT, Rougerie P, Pieuchot L. Role of the nucleus as a sensor of cell environment topography. Adv Healthc Mater 2018; 7:1701154. doi: 10.1002/adhm.201701154 [Crossref] [ Google Scholar]
- Algieri C, Drioli E, Guzzo L, Donato L. Bio-mimetic sensors based on molecularly imprinted membranes. Sensors 2014; 14:13863-912. doi: 10.3390/s140813863 [Crossref] [ Google Scholar]
- Pan J, Chen W, Ma Y, Pan G. Molecularly imprinted polymers as receptor mimics for selective cell recognition. Chem Soc Rev 2018; 47:5574-87. doi: 10.1039/C7CS00854F [Crossref] [ Google Scholar]
- Liu R, Poma A. Advances in Molecularly Imprinted Polymers as Drug Delivery Systems. Molecules 2021; 26. 10.3390/molecules26123589.
- Fresco-Cala B, Batista AD, Cárdenas S. Molecularly Imprinted Polymer Micro- and Nano-Particles. A review. Molecules 2020; 25. 10.3390/molecules25204740.
- Vozzi G, Morelli I, Vozzi F, Andreoni C, Salsedo E, Morachioli A. SOFT‐MI: A novel microfabrication technique integrating soft‐lithography and molecular imprinting for tissue engineering applications. Biotechnol Bioeng 2010; 106:804-17. doi: 10.1002/bit.22740 [Crossref] [ Google Scholar]
- Cieplak M, Kutner W. Artificial biosensors: How can molecular imprinting mimic biorecognition?. Trends Biotechnol 2016; 34:922-41. doi: 10.1016/j.tibtech.2016.05.011 [Crossref] [ Google Scholar]
- Chen L, Xu S, Li J. Recent advances in molecular imprinting technology: current status, challenges and highlighted applications. Chem Soc Rev 2011; 40:2922-42. doi: 10.1039/c0cs00084a [Crossref] [ Google Scholar]
- Yan H, Row KH. Characteristic and synthetic approach of molecularly imprinted polymer. Int J Mol Sci 2006; 7:155-78. doi: 10.3390/i7050155 [Crossref] [ Google Scholar]
- Parisi OI, Ruffo M, Puoci F. Molecularly imprinted polymers for selective recognition in regenerative medicine. Nanostruct Biomater Regen Med 2020; p. 141-63. 10.1186/s40824-023-00388-5.
- Bonatti AF, De Maria C, Vozzi G. Molecular imprinting strategies for tissue engineering applications: A review. Polymers 2021; 13:548. doi: 10.3390/polym13040548 [Crossref] [ Google Scholar]
- Zhang Y, Wang Q, Zhao X, Ma Y, Zhang H, Pan G. Molecularly Imprinted Nanomaterials with Stimuli Responsiveness for Applications in Biomedicine. Molecules 2023; 28:918. doi: 10.3390/molecules28030918 [Crossref] [ Google Scholar]
- Morsi SMM, Abd El-Aziz ME, Mohamed HA. Smart polymers as molecular imprinted polymers for recognition of target molecules. Int J Polym Mater 2023; 72:612-35. doi: 10.1080/00914037.2022.2042287 [Crossref] [ Google Scholar]
- Chen W, Tian X, He W, Li J, Feng Y, Pan G. Emerging functional materials based on chemically designed molecular recognition. BMC Mater 2020; 2:1-22. doi: 10.1186/s42833-019-0007-1 [Crossref] [ Google Scholar]
- Bodoki AE, Iacob B-C, Bodoki E. Perspectives of Molecularly Imprinted Polymer-Based Drug Delivery Systems in Cancer Therapy. Polymers 2019; 11:2085. doi: 10.3390/polym11122085 [Crossref] [ Google Scholar]
- Zhang Y, Deng C, Liu S, Wu J, Chen Z, Li C. Active targeting of tumors through conformational epitope imprinting. Angew Chem Int Ed 2015; 54:5157-60. doi: 10.1002/anie.201412114 [Crossref] [ Google Scholar]
- Liu R, Cui Q, Wang C, Wang X, Yang Y, Li L. Preparation of sialic acid-imprinted fluorescent conjugated nanoparticles and their application for targeted cancer cell imaging. ACS Appl Mater Interfaces 2017; 9:3006-15. doi: 10.1021/acsami.6b14320 [Crossref] [ Google Scholar]
- Pan G, Shinde S, Yeung SY, Jakštaitė M, Li Q, Wingren AG. An Epitope‐Imprinted Biointerface with Dynamic Bioactivity for Modulating Cell–Biomaterial Interactions. Angew Chem Int Ed 2017; 56:15959-63. doi: 10.1002/anie.201708635 [Crossref] [ Google Scholar]
- Kellens E, Bové H, Conradi M, D’Olieslaeger L, Wagner P, Landfester K. Improved Molecular Imprinting Based on Colloidal Particles Made from Miniemulsion: A Case Study on Testosterone and Its Structural Analogues. Macromolecules 2016; 49:2559-67. doi: 10.1021/acs.macromol.6b00130 [Crossref] [ Google Scholar]
- Ding X, Heiden P. Recent Developments in Molecularly Imprinted Nanoparticles by Surface Imprinting Techniques. Macromol Mater Eng 2014; 299. 10.1002/mame.201300160.
- Ren XH, Wang HY, Li S, He XW, Li WY, Zhang YK. Preparation of glycan-oriented imprinted polymer coating Gd-doped silicon nanoparticles for targeting cancer Tn antigens and dual-modal cell imaging via boronate-affinity surface imprinting. Talanta 2021; 223:121706. doi: 10.1016/j.talanta.2020.121706 [Crossref] [ Google Scholar]
- Qin YT, Ma YJ, Feng YS, He XW, Li WY, Zhang YK. Targeted Mitochondrial Fluorescence Imaging-Guided Tumor Antimetabolic Therapy with the Imprinted Polymer Nanomedicine Capable of Specifically Recognizing Dihydrofolate Reductase. ACS Appl Mater Interfaces 2021; 13:40332-41. doi: 10.1021/acsami.1c11388 [Crossref] [ Google Scholar]
- Zaidi S. An Overview of Bio-Inspired Intelligent Imprinted Polymers for Virus Determination. Biosens 2021; 11:89. doi: 10.3390/bios11030089 [Crossref] [ Google Scholar]
- Bodoki AE, Iacob BC, Dinte E, Vostinaru O, Samoila O, Bodoki E. Perspectives of Molecularly Imprinted Polymer-Based Drug Delivery Systems in Ocular Therapy. Polymers (Basel) 2021; 13:3649. doi: 10.3390/polym13213649 [Crossref] [ Google Scholar]
- Jantarat C, Attakitmongkol K, Nichsapa S, Sirathanarun P, Srivaro S. Molecularly imprinted bacterial cellulose for sustained-release delivery of quercetin. J Biomater Sci Polym Ed 2020; 31:1961-76. doi: 10.1080/09205063.2020.1787602 [Crossref] [ Google Scholar]
- Neamtu I, Rusu AG, Diaconu A, Nita LE, Chiriac AP. Basic concepts and recent advances in nanogels as carriers for medical applications. Drug Deliv 2017; 24:539-57. doi: 10.1080/10717544.2016.1276232 [Crossref] [ Google Scholar]
- Zeng Z, Hoshino Y, Rodriguez A, Yoo H, Shea KJ. Synthetic Polymer Nanoparticles with Antibody-like Affinity for a Hydrophilic Peptide. ACS Nano 2010; 4:199-204. doi: 10.1021/nn901256s [Crossref] [ Google Scholar]
- Teixeira SPB, Reis RL, Peppas NA, Gomes ME, Domingues RMA. Epitope-imprinted polymers: Design principles of synthetic binding partners for natural biomacromolecules. Sci Adv 2021; 7:eabi9884. doi: 10.1126/sciadv.abi9884 [Crossref] [ Google Scholar]
- Arabi M, Ostovan A, Li J, Wang X, Zhang Z, Choo J. Molecular Imprinting: Green Perspectives and Strategies. Adv Mater 2021; 33:e2100543. doi: 10.1002/adma.202100543 [Crossref] [ Google Scholar]
- Fu J, Chuah YJ, Liu J, Tan SY, Wang D-A. Respective Effects of Gelatin-Coated Polydimethylsiloxane (PDMS) Substrates on Self-renewal and Cardiac Differentiation of Induced Pluripotent Stem Cells (iPSCs). ACS Biomater Sci Eng 2018; 4:4321-30. doi: 10.1021/acsbiomaterials.8b00993 [Crossref] [ Google Scholar]
- Chuah YJ, Zhang Y, Wu Y, Menon NV, Goh GH, Lee AC. Combinatorial effect of substratum properties on mesenchymal stem cell sheet engineering and subsequent multi-lineage differentiation. Acta Biomater 2015; 23:52-62. doi: 10.1016/j.actbio.2015.05.023 [Crossref] [ Google Scholar]
- Kong HJ, Boontheekul T, Mooney DJ. Quantifying the relation between adhesion ligand–receptor bond formation and cell phenotype. PNAS 2006; 103:18534-9. doi: 10.1073/pnas.0605960103 [Crossref] [ Google Scholar]
- Mashinchian O, Turner L-A, Dalby MJ, Laurent S, Shokrgozar MA, Bonakdar S. Regulation of stem cell fate by nanomaterial substrates. Nanomed 2015; 10:829-47. doi: 10.2217/nnm.14.225 [Crossref] [ Google Scholar]
- Mashinchian O, Bonakdar S, Taghinejad H, Satarifard V, Heidari M, Majidi M. Cell-imprinted substrates act as an artificial niche for skin regeneration. ACS Appl Mater Interfaces 2014; 6:13280-92. doi: 10.1021/am503045b [Crossref] [ Google Scholar]
- Vasapollo G, Sole RD, Mergola L, Lazzoi MR, Scardino A, Scorrano S. Molecularly imprinted polymers: present and future prospective. Int J Mol Sci 2011; 12:5908-45. doi: 10.3390/ijms12095908 [Crossref] [ Google Scholar]
- Kim TK, Kim JK, Jeong OC. Measurement of nonlinear mechanical properties of PDMS elastomer. Microelectron Eng 2011; 88:1982-5. doi: 10.1016/j.mee.2010.12.108 [Crossref] [ Google Scholar]
- Xue P, Li Q, Sun L, Zhang L, Xu Z, Li CM. A simple technique of constructing nano-roughened polydimethylsiloxane surface to enhance mesenchymal stem cell adhesion and proliferation. Microfluid Nanofluidics 2018; 22:1. doi: 10.1007/s10404-017-2014-4 [Crossref] [ Google Scholar]
- Lowry ER, Henderson CE. Stem cell differentiation: yielding substrates for neurons. Nat Mater 2014; 13:543-4. doi: 10.1038/nmat3992 [Crossref] [ Google Scholar]
- Kamguyan K, Katbab AA, Mahmoudi M, Thormann E, Zajforoushan Moghaddam S, Moradi L. An engineered cell-imprinted substrate directs osteogenic differentiation in stem cells. Biomater Sci 2017; 6:189-99. doi: 10.1039/c7bm00733g [Crossref] [ Google Scholar]
- Babaei M, Nasernejad B, Sharifikolouei E, Shokrgozar MA, Bonakdar S. Bioactivation of 3D Cell-Imprinted Polydimethylsiloxane Surfaces by Bone Protein Nanocoating for Bone Tissue Engineering. ACS Omega 2022; 7:26353-67. doi: 10.1021/acsomega.2c02206 [Crossref] [ Google Scholar]
- Kavand H, van Lintel H, Bakhshi Sichani S, Bonakdar S, Kavand H, Koohsorkhi J. Cell-imprint surface modification by contact photolithography-based approaches: Direct-cell photolithography and optical soft lithography using PDMS cell imprints. ACS Appl Mater Interfaces 2019; 11:10559-66. doi: 10.1021/acsami.9b00523 [Crossref] [ Google Scholar]
- Guillemette MD, Cui B, Roy E, Gauvin R, Giasson CJ, Esch MB. Surface topography induces 3D self-orientation of cells and extracellular matrix resulting in improved tissue function. Integr Biol 2009; 1:196-204. doi: 10.1039/b820208g [Crossref] [ Google Scholar]
- Cutiongco MF, Goh SH, Aid-Launais R, Le Visage C, Low HY, Yim EK. Planar and tubular patterning of micro and nano-topographies on poly (vinyl alcohol) hydrogel for improved endothelial cell responses. Biomaterials 2016; 84:184-95. doi: 10.1016/j.biomaterials.2016.01.036 [Crossref] [ Google Scholar]
- Teo BKK, Ankam S, Chan LY, Yim EK. Nanotopography/mechanical induction of stem-cell differentiation. Methods Cell Biol 2010; 98:241-94. doi: 10.1016/s0091-679x(10)98011-4 [Crossref] [ Google Scholar]
- Hoffman-Kim D, Mitchel JA, Bellamkonda RV. Topography, cell response, and nerve regeneration. Annu Rev Biomed Eng 2010; 12:203-31. doi: 10.1146/annurev-bioeng-070909-105351 [Crossref] [ Google Scholar]
- Jeon H, Simon Jr CG, Kim G. A mini‐review: cell response to microscale, nanoscale, and hierarchical patterning of surface structure. J Biomed Mater Res Part B: Appl Biomater 2014; 102:1580-94. doi: 10.1002/jbm.b.33158 [Crossref] [ Google Scholar]
- Nguyen AT, Sathe SR, Yim EK. From nano to micro: topographical scale and its impact on cell adhesion, morphology and contact guidance. J Phys Condens Matter 2016; 28:183001. doi: 10.1088/0953-8984/28/18/183001 [Crossref] [ Google Scholar]
- Washburn NR, Yamada KM, Simon Jr CG, Kennedy SB, Amis EJ. High-throughput investigation of osteoblast response to polymer crystallinity: influence of nanometer-scale roughness on proliferation. Biomaterials 2004; 25:1215-24. doi: 10.1016/j.biomaterials.2003.08.043 [Crossref] [ Google Scholar]
- Arnold M, Cavalcanti‐Adam EA, Glass R, Blümmel J, Eck W, Kantlehner M. Activation of integrin function by nanopatterned adhesive interfaces. Chem Phys Chem 2004; 5:383-8. doi: 10.1002/cphc.200301014 [Crossref] [ Google Scholar]
- Teixeira AI, McKie GA, Foley JD, Bertics PJ, Nealey PF, Murphy CJ. The effect of environmental factors on the response of human corneal epithelial cells to nanoscale substrate topography. Biomaterials2006. 27: 3945-54. 10.1016/j.biomaterials.2006.01.044.
- Leung CM, De Haan P, Ronaldson-Bouchard K, Kim G-A, Ko J, Rho HS. A guide to the organ-on-a-chip. NRMP 2022; 2:1-29. doi: 10.1038/s43586-022-00118-6 [Crossref] [ Google Scholar]
- Pradeep A, Raveendran J, Babu TS. Design, fabrication and assembly of lab-on-a-chip and its uses. PMBTS 2021; 187:121-62. doi: 10.1016/bs.pmbts.2021.07.021 [Crossref] [ Google Scholar]
- Lerman MJ, Lembong J, Muramoto S, Gillen G, Fisher JP. The evolution of polystyrene as a cell culture material. Tissue Eng Part B: Rev 2018; 24:359-72. doi: 10.1089/ten.teb.2018.0056 [Crossref] [ Google Scholar]
- Kapałczyńska M, Kolenda T, Przybyła W, Zajączkowska M, Teresiak A, Filas V. 2D and 3D cell cultures–a comparison of different types of cancer cell cultures. Arch Med Sci 2018; 14:910-9. doi: 10.5114/2Faoms.2016.63743 [Crossref] [ Google Scholar]
- Lee KY, Mooney DJ. Alginate: properties and biomedical applications. Prog Polym Sci 2012; 37:106-26. doi: 10.1016/j.progpolymsci.2011.06.003 [Crossref] [ Google Scholar]
- Gheorghita Puscaselu R, Lobiuc A, Dimian M, Covasa M. Alginate: From food industry to biomedical applications and management of metabolic disorders. Polymers 2020; 12:2417. doi: 10.3390/polym12102417 [Crossref] [ Google Scholar]
- Shin MJ, Shin YJ, Shin JS. Sensing capability of molecularly imprinted self-assembled monolayer fabricated using dithiol compound. J Ind Eng Chem 2014; 20:91-5. doi: 10.1016/j.jiec.2013.04.022 [Crossref] [ Google Scholar]
- Gunti S, Hoke AT, Vu KP, London Jr NR. Organoid and spheroid tumor models: Techniques and applications. Cancers 2021; 13:874. doi: 10.3390/2Fcancers13040874 [Crossref] [ Google Scholar]
- Abu-Hakmeh AE, Wan LQ. High-throughput cell aggregate culture for stem cell chondrogenesis. Biomim Stem Cells 2014; 4:11-9. doi: 10.1007/7651_2014_75 [Crossref] [ Google Scholar]
- Chung JJ, Im H, Kim SH, Park JW, Jung Y. Toward biomimetic scaffolds for tissue engineering: 3D printing techniques in regenerative medicine. Front Bioengin Biotechnol 2020; 8:586406. doi: 10.3389/fbioe.2020.586406 [Crossref] [ Google Scholar]
- Huang C, Ozdemir T, Xu LC, Butler PJ, Siedlecki CA, Brown JL. The role of substrate topography on the cellular uptake of nanoparticles. J Biomed Mater Res B Appl Biomater 2016; 104:488-95. doi: 10.1002/jbm.b.33397 [Crossref] [ Google Scholar]
- Adhikari J, Roy A, Chanda A, Gouripriya D, Thomas S, Ghosh M. Effects of surface patterning and topography on cellular functions of tissue engineered scaffolds with special reference to 3D bioprinting. Biomater Sci 2023; 11:1236-1269. doi: 10.1039/D2BM01499H [Crossref] [ Google Scholar]
- Tan LH, Sykes PH, Alkaisi MM, Evans JJ. The characteristics of Ishikawa endometrial cancer cells are modified by substrate topography with cell-like features and the polymer surface. Int J Nanomedicine 2015; 4883-95. 10.2147/ijn.s86336.
- Domura R, Sasaki R, Ishikawa Y, Okamoto M. Cellular morphology-mediated proliferation and drug sensitivity of breast cancer cells. J Func Biomater 2017; 8:18. doi: 10.3390/jfb8020018 [Crossref] [ Google Scholar]
- Tan LH, Sykes PH, Alkaisi MM, Evans JJ. Cell-like features imprinted in the physical nano-and micro-topography of the environment modify the responses to anti-cancer drugs of endometrial cancer cells. Biofabrication 2017; 9:015017. doi: 10.1088/1758-5090/aa5c9a [Crossref] [ Google Scholar]
- Kim HN, Jiao A, Hwang NS, Kim MS, Kim D-H, Suh K-Y. Nanotopography-guided tissue engineering and regenerative medicine. Adv Drug Deliv Rev 2013; 65:536-58. doi: 10.1016/j.addr.2012.07.014 [Crossref] [ Google Scholar]
- Kamguyan K, Moghaddam SZ, Nazbar A, Haramshahi SMA, Taheri S, Bonakdar S. Cell-imprinted substrates: in search of nanotopographical fingerprints that guide stem cell differentiation. Nanoscale Adv 2021; 3:333-8. doi: 10.1039/2Fd0na00692k [Crossref] [ Google Scholar]
- Von Der Mark K, Gauss V, Von Der Mark H, Müller P. Relationship between cell shape and type of collagen synthesised as chondrocytes lose their cartilage phenotype in culture. Nature 1977; 267:531-2. doi: 10.1038/267531a0 [Crossref] [ Google Scholar]
- Hoffman T, Khademhosseini A, Langer R. Chasing the paradigm: clinical translation of 25 years of tissue engineering. Tissue Eng Part A 2019; 25:679-87. doi: 10.1089/ten.tea.2019.0032 [Crossref] [ Google Scholar]
- Bonakdar S, Mahmoudi M, Montazeri L, Taghipoor M, Bertsch A, Shokrgozar MA. Cell-imprinted substrates modulate differentiation, redifferentiation, and transdifferentiation. ACS Appl Mater Interfaces 2016; 8:13777-84. doi: 10.1021/acsami.6b03302 [Crossref] [ Google Scholar]
- Dalby MJ, Gadegaard N, Oreffo RO. Harnessing nanotopography and integrin–matrix interactions to influence stem cell fate. Nat Mater 2014; 13:558-69. doi: 10.1038/nmat3980 [Crossref] [ Google Scholar]
- Freyria A-M, Mallein-Gerin F. Chondrocytes or adult stem cells for cartilage repair: the indisputable role of growth factors. Injury 2012; 43:259-65. doi: 10.1016/j.injury.2011.05.035 [Crossref] [ Google Scholar]
- Watt FM. Effect of seeding density on stability of the differentiated phenotype of pig articular chondrocytes in culture. J Cell Sci 1988; 89:373-8. doi: 10.1242/jcs.89.3.373 [Crossref] [ Google Scholar]
- Hong E, Reddi AH. Dedifferentiation and redifferentiation of articular chondrocytes from surface and middle zones: changes in microRNAs-221/-222,-140, and-143/145 expression. Tissue Eng Part A 2013; 19:1015-22. [ Google Scholar]
- Darling EM, Athanasiou KA. Rapid phenotypic changes in passaged articular chondrocyte subpopulations. J Orthop Res 2005; 23:425-32. doi: 10.1016/j.orthres.2004.08.008 [Crossref] [ Google Scholar]
- Kamguyan K, Katbab AA, Mahmoudi M, Thormann E, Moghaddam SZ, Moradi L. An engineered cell-imprinted substrate directs osteogenic differentiation in stem cells. Biomater Sci 2018; 6:189-99. doi: 10.1039/C7BM00733G [Crossref] [ Google Scholar]
- Kamguyan K, Zajforoushan Moghaddam S, Nazbar A, Haramshahi SMA, Taheri S, Bonakdar S. Cell-imprinted substrates: in search of nanotopographical fingerprints that guide stem cell differentiation. Nanoscale Adv 2021; 3:333-338. doi: 10.1039/D0NA00692K [Crossref] [ Google Scholar]
- Alam SG, Zhang Q, Prasad N, Li Y, Chamala S, Kuchibhotla R. The mammalian LINC complex regulates genome transcriptional responses to substrate rigidity. Sci Rep 2016; 6:38063. doi: 10.1038/srep38063 [Crossref] [ Google Scholar]
- Haramshahi SMA, Bonakdar S, Moghtadaei M, Kamguyan K, Thormann E, Tanbakooei S. Tenocyte-imprinted substrate: a topography-based inducer for tenogenic differentiation in adipose tissue-derived mesenchymal stem cells. Biomed Mater 2020; 15:035014. doi: 10.1088/1748-605x/ab6709 [Crossref] [ Google Scholar]
- Ghazali ZS, Eskandari M, Bonakdar S, Renaud P, Mashinchian O, Shalileh S. Neural priming of adipose-derived stem cells by cell-imprinted substrates. Biofabrication 2020; 13:1-22. doi: 10.1088/1758-5090/abc66f [Crossref] [ Google Scholar]
- Lee EA, Im SG, Hwang NS. Efficient myogenic commitment of human mesenchymal stem cells on biomimetic materials replicating myoblast topography. Biotechnol J 2014; 9:1604-12. doi: 10.1002/biot.201400020 [Crossref] [ Google Scholar]
- Moosazadeh Moghaddam M, Bonakdar S, Shokrgozar MA, Zaminy A, Vali H, Faghihi S. Engineered substrates with imprinted cell-like topographies induce direct differentiation of adipose-derived mesenchymal stem cells into Schwann cells. Artif Cells Nanomed Biotechnol 2019; 47:1022-35. doi: 10.1080/21691401.2019.1586718 [Crossref] [ Google Scholar]
- Abadi PP, Garbern JC, Behzadi S, Hill MJ, Tresback JS, Heydari T. Engineering of mature human induced pluripotent stem cell‐derived cardiomyocytes using substrates with multiscale topography. Adv Func Mater 2018; 28:1-18. doi: 10.1002/adfm.201870128 [Crossref] [ Google Scholar]
- Jeon H, Kim G. Effects of a cell-imprinted poly (dimethylsiloxane) surface on the cellular activities of MG63 osteoblast-like cells: preparation of a patterned surface, surface characterization, and bone mineralization. Langmuir 2012; 28:13423-30. doi: 10.1021/la302937k [Crossref] [ Google Scholar]
- Wang W, Cui H, Zhang P, Meng J, Zhang F, Wang S. Efficient capture of cancer cells by their replicated surfaces reveals multiscale topographic interactions coupled with molecular recognition. ACS Appl Mater Interfaces 2017; 9:10537-43. doi: 10.1021/acsami.7b01147 [Crossref] [ Google Scholar]
- Farvadi F, Ghahremani MH, Hashemi F, Hormozi-Nezhad MR, Raoufi M, Zanganeh S. Cell shape affects nanoparticle uptake and toxicity: An overlooked factor at the nanobio interfaces. J Colloid Interface Sci 2018; 531:245-52. doi: 10.1016/j.jcis.2018.07.013 [Crossref] [ Google Scholar]
- Kavand H, Rahaie M, Koohsorkhi J, Haghighipour N, Bonakdar S. A conductive cell‐imprinted substrate based on CNT–PDMS composite. Biotechnol Appl Biochem 2019; 66:445-53. doi: 10.1002/bab.1741 [Crossref] [ Google Scholar]
- Gholami H, Mardjanmehr SH, Dehghan MM, Bonakdar S, Farzad Mohajeri S. Osteogenic Differentiation of Mesenchymal Stem Cells Via Osteoblast-Imprinted Substrate: In Vitro and In Vivo Evaluation in Rat Model. Iran J Vet Med 2019; 13:260-9. [ Google Scholar]
- Haramshahi SMA, Bonakdar S, Moghtadaei M, Kamguyan K, Thormann E, Tanbakooei S. Tenocyte-imprinted substrate: a topography-based inducer for tenogenic differentiation in adipose tissue-derived mesenchymal stem cells. Biomed Mater 2020; 15:035014. doi: 10.1088/1748-605X/ab6709 [Crossref] [ Google Scholar]
- Kashani SY, Moraveji MK, Taghipoor M, Kowsari-Esfahan R, Hosseini AA, Montazeri L. An integrated microfluidic device for stem cell differentiation based on cell-imprinted substrate designed for cartilage regeneration in a rabbit model. Mater Sci Eng C 2020; 111794:1-12. doi: 10.1016/j.msec.2020.111794 [Crossref] [ Google Scholar]
- Yazdian Kashani S, Keshavarz Moraveji M, Bonakdar S. Computational and experimental studies of a cell-imprinted-based integrated microfluidic device for biomedical applications. Sci Rep 2021; 11:12130. doi: 10.1038/s41598-021-91616-2 [Crossref] [ Google Scholar]
- Dadashkhan S, Irani S, Bonakdar S, Ghalandari B. P75 and S100 gene expression induced by cell‐imprinted substrate and beta‐carotene to nerve tissue engineering. J Appl Polymer Sci 2021; 138:50624. doi: 10.1002/app.50624 [Crossref] [ Google Scholar]
- Nazbar A, Samani S, Yazdian Kashani S, Amanzadeh A, Shoeibi S, Bonakdar S. Molecular imprinting as a simple way for the long-term maintenance of the stemness and proliferation potential of adipose-derived stem cells: an in vitro study. J Mater Chem B 2022; 10:6816-30. doi: 10.1039/D2TB00279E [Crossref] [ Google Scholar]
- Chung C, Burdick JA. Engineering cartilage tissue. Adv Drug Deliv Rev 2008; 60:243-62. doi: 10.1016/j.addr.2007.08.027 [Crossref] [ Google Scholar]
- Zhao W, Wang T, Luo Q, Chen Y, Leung VY, Wen C. Cartilage degeneration and excessive subchondral bone formation in spontaneous osteoarthritis involves altered TGF‐β signaling. J Orthop Res 2016; 34:763-70. doi: 10.1002/jor.23079 [Crossref] [ Google Scholar]
- Hubka KM, Dahlin RL, Meretoja VV, Kasper FK, Mikos AG. Enhancing chondrogenic phenotype for cartilage tissue engineering: monoculture and coculture of articular chondrocytes and mesenchymal stem cells. Rev 2014; 20:Rev 2014; 20. doi: 10.1089/ten.teb.2014.0034 [Crossref] [ Google Scholar]
- Ohashi K, Fujiwara S, Mizuno K. Roles of the cytoskeleton, cell adhesion and rho signalling in mechanosensing and mechanotransduction. J Biochem 2017; 161:245-54. doi: 10.1093/jb/mvw082 [Crossref] [ Google Scholar]
- Martino F, Perestrelo AR, Vinarský V, Pagliari S, Forte G. Cellular mechanotransduction: from tension to function. Front Physiol 2018; 9:824. doi: 10.3389/fphys.2018.00824 [Crossref] [ Google Scholar]
- Harris AR, Jreij P, Fletcher DA. Mechanotransduction by the actin cytoskeleton: converting mechanical stimuli into biochemical signals. Annu Rev Biophys 2018; 47:617-31. doi: 10.1146/annurev-biophys-070816-033547 [Crossref] [ Google Scholar]
- Hernando JB, Gómez MR, Lantada AD. Modeling Living cells Within Microfluidic Systems Using Cellular Automata Models. Sci Rep 2019; 9:1-10. doi: 10.1038/s41598-019-51494-1 [Crossref] [ Google Scholar]
- Lund AW, Yener B, Stegemann JP, Plopper GE. The natural and engineered 3D microenvironment as a regulatory cue during stem cell fate determination. Tissue Eng Part B: Rev 2009; 15:371-80. doi: 10.1089/ten.teb.2009.0270 [Crossref] [ Google Scholar]
- Cun X, Hosta-Rigau L. Topography: a biophysical approach to direct the fate of mesenchymal stem cells in tissue engineering applications. Nanomater 2020; 10:2070. doi: 10.3390/nano10102070 [Crossref] [ Google Scholar]
- Ravichandran R, Liao S, Ng CC, Chan CK, Raghunath M, Ramakrishna S. Effects of nanotopography on stem cell phenotypes. WJSC 2009; 1:55. doi: 10.4252/wjsc.v1.i1.55 [Crossref] [ Google Scholar]
- Khanal G, Chung K, Solis-Wever X, Johnson B, Pappas D. Ischemia/reperfusion injury of primary porcine cardiomyocytes in a low-shear microfluidic culture and analysis device. Analyst 2011; 136:3519-26. doi: 10.1039/c0an00845a [Crossref] [ Google Scholar]
- Wood DK, Soriano A, Mahadevan L, Higgins JM, Bhatia SN. A biophysical indicator of vaso-occlusive risk in sickle cell disease. Sci Transl Med 2012; 4:123ra26-ra26. doi: 10.1126/scitranslmed.3002738 [Crossref] [ Google Scholar]
- Jang K-J, Suh K-Y. A multi-layer microfluidic device for efficient culture and analysis of renal tubular cells. Lab Chip 2010; 10:36-42. doi: 10.1039/b907515a [Crossref] [ Google Scholar]
- Snouber LC, Letourneur F, Chafey P, Broussard C, Monge M, Legallais C. Analysis of transcriptomic and proteomic profiles demonstrates improved Madin–Darby canine kidney cell function in a renal microfluidic biochip. Biotechnol Prog 2012; 28:474-84. doi: 10.1002/btpr.743 [Crossref] [ Google Scholar]
- Jang K-J, Mehr AP, Hamilton GA, McPartlin LA, Chung S, Suh K-Y. Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integ Biol 2013; 5:1119-29. doi: 10.1039/c3ib40049b [Crossref] [ Google Scholar]
- Hoes MF, Bomer N, van der Meer P. Concise review: the current state of human in vitro cardiac disease modeling: A focus on gene editing and tissue engineering. Stem Cells Transl Med 2019; 8:66-74. doi: 10.1002/sctm.18-0052 [Crossref] [ Google Scholar]
- Boland MJ, Hazen JL, Nazor KL, Rodriguez AR, Gifford W, Martin G. Adult mice generated from induced pluripotent stem cells. Nature 2009; 461:91-4. doi: 10.1038/nature08310 [Crossref] [ Google Scholar]
- Shi Y, Inoue H, Wu JC, Yamanaka S. Induced pluripotent stem cell technology: a decade of progress. Nat Rev Drug Discov 2017; 16:115. doi: 10.1038/nrd.2016.245 [Crossref] [ Google Scholar]
- Liu C, Oikonomopoulos A, Sayed N, Wu JC. Modeling human diseases with induced pluripotent stem cells: from 2D to 3D and beyond. Development 2018; 145:dev156166. doi: 10.1242/dev.156166 [Crossref] [ Google Scholar]
- McCauley HA, Wells JM. Pluripotent stem cell-derived organoids: using principles of developmental biology to grow human tissues in a dish. Development 2017; 144:958-62. doi: 10.1242/dev.140731 [Crossref] [ Google Scholar]
- Quadrato G, Brown J, Arlotta P. The promises and challenges of human brain organoids as models of neuropsychiatric disease. Nat Med 2016; 22:1220. doi: 10.1038/nm.4214 [Crossref] [ Google Scholar]
- McCracken KW, Catá EM, Crawford CM, Sinagoga KL, Schumacher M, Rockich BE. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 2014; 516:400-4. doi: 10.1038/nature13863 [Crossref] [ Google Scholar]
- Murfee WL, Sweat RS, Tsubota K-i, Gabhann FM, Khismatullin D, Peirce SM. Applications of computational models to better understand microvascular remodelling: a focus on biomechanical integration across scales. Interface Focus 2015; 5:20140077. doi: 10.1098/rsfs.2014.0077 [Crossref] [ Google Scholar]
- Jain K. Cell therapy: technologies, markets and companies. Basel: Jain PharmaBiotech 2013.
- Arjmand B, Goodarzi P, Mohamadi-Jahani F, Falahzadeh K, Larijani B. Personalized regenerative medicine. Acta Med Iran 2017; 144-9.
- Kim I. A brief overview of cell therapy and its product. JKAOMS 2013; 39:201-2. doi: 10.5125/jkaoms.2013.39.5.201 [Crossref] [ Google Scholar]
- Chu D-T, Nguyen TT, Tien NLB, Tran D-K, Jeong J-H, Anh PG. Recent Progress of Stem Cell Therapy in Cancer Treatment: Molecular Mechanisms and Potential Applications. Cells 2020; 9:563. doi: 10.3390/cells9030563 [Crossref] [ Google Scholar]
- Perin EC. Stem cell therapy for cardiovascular disease. Texas Heart Institute Journal 2006; 33:204. [ Google Scholar]
- Kobayashi N. Cell therapy for diabetes mellitus. Cell Transplant 2006; 15:849-54. doi: 10.3727/000000006783981396 [Crossref] [ Google Scholar]
- Counsel PD, Bates D, Boyd R, Connell DA. Cell therapy in joint disorders. Sports Health 2015; 7:27-37. doi: 10.1177/1941738114523387 [Crossref] [ Google Scholar]
- Steward C, Jarisch A. Haemopoietic stem cell transplantation for genetic disorders. ADC 2005; 90:1259-63. doi: 10.1136/adc.2005.074278 [Crossref] [ Google Scholar]
- Dehkordi AN, Babaheydari FM, Chehelgerdi M, Dehkordi SR. Skin tissue engineering: wound healing based on stem-cell-based therapeutic strategies. Stem Cell Res Ther 2019; 10:111. [ Google Scholar]
- Sánchez A, Schimmang T, García-Sancho J. Cell and tissue therapy in regenerative medicine. Stem Cell Transplant 2012. p. 89-102. 10.1007/978-1-4614-2098-9_7.
- Mousavinejad M, Andrews PW, Shoraki EK. Current biosafety considerations in stem cell therapy. Cell J (Yakhteh) 2016; 18:281. doi: 10.22074/cellj.2016.4324 [Crossref] [ Google Scholar]
- Moradi L, Vasei M, Dehghan MM, Majidi M, Mohajeri SF, Bonakdar S. Regeneration of meniscus tissue using adipose mesenchymal stem cells-chondrocytes co-culture on a hybrid scaffold: In vivo study. Biomaterials 2017; 126:18-30. doi: 10.1016/j.biomaterials.2017.02.022 [Crossref] [ Google Scholar]