Bioimpacts. 2025;15:30688.
doi: 10.34172/bi.30688
Original Article
Chrysosplenetin B suppresses the growth of human prostate cancer cells by inducing G1 cell cycle arrest
Gang He Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing, 1, 2, , * # 
Yanjiao Feng Data curation, Formal analysis, Investigation, Methodology, Resources, Validation, Visualization, Writing – original draft, 1, #
Tangcong Chen Data curation, Investigation, Methodology, Resources, 1
Yiyuan Zhang Methodology, Resources, 1
Li Liang Data curation, Investigation, 1
Jun Yan Investigation, 1
Yanxia Song Writing – review & editing, 2
Fengzheng Chen Writing – review & editing, 3
Wei Liu Formal analysis, Funding acquisition, Writing – review & editing, 1, 4, * 
Author information:
1Key Laboratory of Medicinal and Edible Plants Resources Development of Sichuan Education Department, Sichuan Industrial Institute of Antibiotics, School of Pharmacy, Chengdu University, Chengdu, China
2Antibiotics Research and Re-evaluation Key Laboratory of Sichuan Province, Sichuan Industrial Institute of Antibiotics, School of Pharmacy, Chengdu University, Chengdu, China
3Sichuan Province Key Laboratory of Natural Products and Small Molecule Synthesis, Leshan Normal University, Leshan, China
4School of Life Science, Leshan Normal University, Leshan, China
#These authors contributed equally to this work.
Abstract
Introduction:
Prostate cancer (PCa) often progresses to castration-resistant prostate cancer (CRPC), which is linked to higher treatment resistance and recurrence rates. This highlights the urgent need for new therapeutic options. Natural products, especially flavonoids, have shown promise in reducing drug resistance and possess both antioxidant and anticancer effects. Developing drugs that specifically target CRPC could offer significant therapeutic advantages.
Methods:
Chrysosplenetin B (CspB) was extracted and purified from the herb Laggera pterodonta (DC.) Benth. using traditional flavonoid extraction techniques, followed by high-performance liquid chromatography (HPLC) for purity assessment and nuclear magnetic resonance (NMR) for structural identification. The effect of CspB on the viability of PCa cells was evaluated using the Cell Counting Kit-8 assay. Subsequently, transcriptome analysis was conducted, and cell cycle progression was assessed through flow cytometry in conjunction with propidium iodide (PI) staining. Additionally, western blotting and quantitative real-time polymerase chain reaction (qRT-PCR) were employed to confirm the expression levels of relevant proteins and genes.
Results:
CspB was found to inhibit the proliferation of PC3, DU145, and LNCaP cells in a dose-dependent manner, with a stronger effect noted in PC3 and DU145 cells. Transcriptomic analysis revealed that CspB treatment led to cell cycle arrest, particularly in PC3 cells. Flow cytometry with PI staining confirmed that CspB caused G1 phase cell cycle arrest in PC3 cells. Moreover, CspB treatment significantly increased the expression of essential members of the Cip/Kip family, including CIP1/P21 and KIP1/P27, as well as CDKN2B (P15) and CDKN2D (P19) from the INK4 family. Additionally, CspB exposure notably raised the expression of the G1 phase-negative regulatory gene CDKN1C, while key cell cycle regulators like CDK6 and E2F1 were significantly downregulated at the protein level.
Conclusion:
Our findings indicate that CspB effectively inhibits the proliferation of CRPC cells by reducing the activity of cell cycle proteins and cyclin-dependent kinase (CDK) complexes while upregulating the expression of P21 and P27 and inducing G1 phase cell cycle arrest. These results highlight the potential of CspB as a promising candidate for developing therapeutic agents aimed at targeting CRPC.
Keywords: Chrysosplenetin B, Suppress cell viability, Prostate cancer, Cell cycle arrest
Copyright and License Information
© 2025 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Funding Statement
This work was supported by the Sichuan Science and Technology Program (Grant Number: 2019YFH0054 and 2020YFH0205), Opening Project of Sichuan Province Key Laboratory of Natural Products and Small Molecule Synthesis (Grant Number: TRCWYXFZCH2022B01and TRCWYXFZCH2022B02) and the Personnel Training Quality and Teaching Reform Project of Chengdu University (Grant Number: CDJGB2022156).
Introduction
Prostate cancer (PCa) ranks as the second most prevalent cancer type and is the fifth leading cause of cancer-related mortality among men globally, with approximately 40 000 deaths reported annually.1 The incidence and mortality rates of PCa are markedly higher in developed nations compared to developing countries, a disparity that may be attributed to various factors, including dietary habits, ethnicity, age, and genetic mutations.2 Androgen receptor (AR) signal transduction is integral to the pathogenesis of PCa, and androgen deprivation therapy (ADT) has been established as the standard first-line treatment for patients with advanced disease.3 Typically, effective management of localized PCa involves either active surveillance or local interventions, such as radical prostatectomy and radiotherapy. For a minority of patients with metastatic PCa, ADT has emerged as the primary therapeutic approach. However, most patients develop tolerance to ADT, which is linked to diverse biological responses and ultimately leads to the emergence of castration-resistant prostate cancer (CRPC).4 The past decade has approved several effective AR-targeted therapies, including abiraterone,5 enzalutamide,6 apalutamide,7 and darolutamide,8 which have significantly advanced the treatment landscape for CRPC. The mechanism of action of AR inhibitors involves their binding to the ligand-binding domain (LBD) encoded by exons 5-6 of the AR gene, thereby obstructing the interaction between androgens and the LBD, which results in a therapeutic effect.9 Notably, approximately 60% of patients with metastatic CRPC exhibit mutations and amplifications in the AR gene, which diminish the binding affinity of AR inhibitors to the LBD and progressively contribute to developing resistance.10 Therefore, identifying and developing novel bioactive compounds are critical for advancing PCa therapies. Additionally, multiple signalling pathways, including PTEN/PI3K/AKT/mTOR, DNA repair mechanisms, and cell cycle regulation, interact with AR signalling in PCa pathogenesis and progression. Consequently, targeting these interconnected pathways represents a promising strategy for developing new therapeutic agents.
The active components of traditional Chinese medicines present a promising avenue for developing novel pharmaceuticals. Numerous studies have indicated that the bioactive constituents extracted from these traditional remedies can modulate the expression of genes associated with cyclins, cyclin-dependent kinases (CDKs), and CDK inhibitors. This modulation plays a critical role in regulating the survival and apoptosis of PCa cells, while simultaneously inhibiting their uncontrolled proliferation. One notable natural anticancer agent, amygdalin, which is derived from bitter almonds, has been investigated for its effects on human LNCaP PCa cells and DU-145 and PC3 CRPC cells. Amygdalin has been shown to effectively modulate the activity of CDK1, CDK2, and CDK4, in addition to influencing the expression of cyclin B and cyclin D. As a result, it induces cell cycle arrest at the G0/G1 phase.11 Furthermore, amygdalin has demonstrated inhibitory effects on the growth and proliferation of human bladder cancer cell lines, including UMUC-3, RT112, and TCCSUP, by downregulating the protein levels of CDK2 and cyclin A, which also leads to cell cycle arrest at the G0/G1 phase.12
Chrysosplenetin B (CspB) is a flavonoid isolated from the herb Laggera pterodonta, first documented in the Yunnan Materia Medica as a traditional Chinese medicinal herb. The primary active constituents of Laggera pterodonta include sesquiterpenoids, flavonoids, and volatile oils. As a result, this herb exhibits a range of pharmacological effects, such as anti-inflammatory, analgesic, expectorant, antipyretic, antiviral, and anti-tumour activities, thereby rendering it useful in the prevention and treatment of various conditions, including COVID-19, colds, malaria, and other diseases.13 A total of 23 compounds, comprising sesquiterpenoids and flavonoids, have been isolated and characterized from Laggera pterodonta, with a screening process identifying seven compounds that demonstrate significant anti-inflammatory properties. Furthermore, research conducted by Yu et al has shown that sesquiterpenoids derived from Laggera pterodonta exhibit cytotoxic effects on HepG2 liver cancer cells.14
Patel previously elucidated the diverse pharmacological activities of chrysosplenetin, which include antiproliferative, antibacterial, and anti-inflammatory effects, by systematically organizing and analyzing data from various databases.15 Lan et al extracted a range of flavonoids from Artemisia annua, including chrysosplenetin.16 The investigation into its antibacterial effects against drug-resistant Staphylococcus aureus revealed that chrysosplenetin B, in combination with norfloxacin, exhibited enhanced antibacterial activity against the drug-resistant strain SA1199B. Furthermore, Treatment of HeLa cells with escalating concentrations of CspB resulted in an increased percentage of cells in the G2/M phase. Additionally, CspB was found to enhance apoptosis rates in both HeLa and A549 cell lines. Furthermore, it exhibited selective inhibition of proliferation in breast cancer cell lines, including MDA-MB-231, MCF-7, and T47D, while demonstrating relatively low toxicity towards normal non-tumor cells, such as MRC-5 and HUVEC.17 Notably, the regulation of kinases induced by flavonoids is strongly associated with apoptosis and proliferation18; a diet rich in flavonoids has been shown to reduce the risk of colon, prostate, and breast cancer.19 Hong et al evaluated the effects of CspB on the osteogenesis of human-derived bone marrow stromal cells and its potential to inhibit osteoporosis induced by estrogen deficiency through the Wnt/β-catenin signalling pathway.20 Sinha et al investigated CspB and artemisinin, both isolated from Tanacetum gracile and found that these compounds inhibit proliferation by obstructing PI3K/Akt signal transduction in breast cancer cells, thereby enhancing apoptosis through mitotic arrest.21
This study primarily investigates the inhibitory effects of CspB on three PCa cell lines. The LNCaP cells exhibit early androgen-dependent characteristics, whereas the androgen-independent PC3 and DU145 PCa cells demonstrate significant metastatic potential.22 Consequently, selecting these three cell lines serves as an effective model for replicating the progression and metastasis of PCa in clinical contexts. However, the signalling pathways and molecular mechanisms through which CspB interacts with human prostate cancer remain unidentified. Transcriptome analysis indicated that the differentially expressed genes predominantly influence the cell cycle. We hypothesized that CspB selectively targets critical cell cycle pathways, thereby exerting growth-inhibitory effects in CRPC cell lines. Our findings, presented here for the first time, demonstrate that CspB inhibits the activity of the cell cycle protein/CDK complex and induces the expression of P21 and P27, leading to G1 phase cell cycle arrest. This mechanism may elucidate the suppressive capacity of CspB in regulating cellular proliferation.
Materials and Methods
Materials and reagents
Laggera pterodonta (DC.) Benth herb was purchased from Chengdu’s traditional Chinese medicine market. The CspB standard (purity ≥ 98%) was purchased from Biopurify Phytochemicals, Ltd. (Chengdu, China). RPMI-1640 medium and DMEM (Gibco; Thermo Fisher Scientific, USA) containing 10% fetal bovine serum (Gibco; Thermo Fisher Scientific) and 1% antibiotics (penicillin 10,000 U/mL, streptomycin 10 mg/mL) (Gibco; Thermo Fisher Scientific) were used to culture the cells. Trypsin-EDTA was obtained from Gibco (Thermo Fisher Scientific, Waltham, MA, USA). Primary antibodies against DP1, CDKN2C, CDK6, E2F1, and β-actin were used for western blot analysis and were purchased from HUABIO (Hangzhou, China). The secondary antibodies were purchased from Beyotime Biotech. Inc. (Shanghai, China). All chromatographic and analytical pure chemicals were purchased from Chengdu Kelong Chemical Co., Ltd. (Chengdu, China).
Isolation and identification of CspB
A total of 2 kg crushed Laggera pterodonta (DC.) Benth was immersed in 95% ethanol at room temperature for 5 days, followed by concentration of the liquid supernatant under reduced pressure to obtain the ethanol extract of Laggera pterodonta. The extract was then dispersed in 500 mL of water and subjected to two successive extractions with 500 mL of petroleum each to remove lipophilic components. Subsequently, two additional extractions with 500 mL of ethyl acetate each were conducted, and the resultant extract was concentrated under reduced pressure to obtain the ethyl acetate extract of Laggera pterodonta. The ethyl acetate underwent separation and purification via silica gel column chromatography. The elution conditions for the silica gel column involved a gradient elution with a mixture of petroleum ether and acetone (6:1 to 1:1, v:v).15 The fractions collected from the thin layer were analyzed to detect individual components and prepare the monomeric compound through preparative high-performance liquid chromatography (HPLC) separation and purification.23 A C18 preparative column was employed with methanol-water (60:40) as the mobile phase at a 5 mL/min flow rate and was monitored by a UV detector for preparative HPLC preparation. The monomeric compounds were dissolved in deuterated DMSO and subjected to nuclear magnetic resonance (NMR) analysis using Bruker AV400. HPLC verified chromatographic conditions; column: Themo Hypersil GOLD, 4.6 × 250 mm, 5.0 μm; column temperature: 30 °C; detection mode: UV 257 nm; flow rate: 1.0 mL/min, sample dissolution: methanol; mobile phase: A, acetonitrile and B, 0.1% phosphoric acid in water; gradient elution: A, 35-65% in 20 min and to 85% in 10 min.
Cell lines and culture conditions
The PC3 and LNCaP PCa cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA) and were provided by Dr. Jin Xi from the Urology Institute of Sichuan University. The DU145 cell line was acquired from Chengdu Miracle Technology Inc. The PC3 and LNCaP cells were cultured in RPMI-1640 medium supplemented with 10% (v/v) fetal bovine serum and 1% (v/v) penicillin-streptomycin, while the DU145 cells were maintained in DMEM containing 10% (v/v) fetal bovine serum and 1% (v/v) penicillin-streptomycin. All cell lines were incubated in a cell culture incubator at 37 °C with 5% CO2.24,25
Cell viability assay
The cytotoxicity of CspB was assessed using the Cell Counting Kit (CCK)-8 cell proliferation and cytotoxicity assay kit.26 The cultured cells were inoculated in 96-well plates at 1 × 104 cells/well density. After overnight incubation for cell attachment, all three cell lines were treated with various concentrations (20, 40, 60, 80, 100 and 120 μM) of CspB for different durations (12, 24 and 48 h). The control group was treated with an equal volume of serum-containing medium. Following the respective treatment times, the medium was removed, and the cells were incubated with CCK-8 reagent (10 μL) for 30 min. The absorbance of the cells was measured at 450 nm using a microplate reader. Cell viability (%) relative to untreated cells was calculated as the percentage of viable cells compared to untreated cells. The line graphs were plotted using GraphPad Prism 8.0 (San Diego, CA, USA), and the IC50 values, along with their corresponding 95% confidence intervals, were determined through a non-linear regression analysis of six independent experiments.
Transcriptome sequencing
A transcriptome sequencing analysis was conducted on PC3 cells treated with CspB to explore the molecular mechanisms by which CspB inhibits the survival of prostate cancer cells. This approach aimed to identify the differentially expressed genes and elucidate the main biological functions involved. Total RNA was extracted from the cell samples, and mRNA was enriched after DNA digestion with DNase. Double-stranded cDNA was synthesized by purification and end-repair. An A-tail was added to the 3' end, and the cDNA was ligated into the sequencing junction. Polymerase chain reaction (PCR) was then performed to amplify the enriched fragments. The constructed libraries were assessed using an Agilent 2100 Bioanalyzer, and sequencing was carried out on an Illumina HiSeqTM 2500 platform to generate 125 or 150 base pair (bp) paired-end data. The raw data produced by high-throughput sequencing were filtered and aligned to the reference genome of the species using HISAT2, and the genome matching rate for the samples was evaluated.27
Gene expression values were quantified as FPKM (Fragments Per Kilobase of transcript per Million mapped reads) using cufflink software (https://cole-trapnell-lab.github.io/cufflinks/).28 To assess the expression differences among the genes, the number of reads for each sample was determined using HTSeq-count software,29 Subsequently, the data were normalized utilizing the estimate size factors function from the DESeq (2012) R package. The NbinomTest function was employed to calculate the P-value and fold-change values.30 Genes exhibiting P < 0.05 and multiple differences more significant than two were identified as differentially expressed. Furthermore, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were conducted to elucidate the biological functions and pathways predominantly influenced by the differentially expressed genes. Additionally, unsupervised hierarchical clustering was performed on these genes, and the expression patterns across different samples were visualized through heat maps.
DNA content and cell cycle analysis
Flow cytometry was employed to assess the DNA content and cell cycle distribution of three PCa cell lines subjected to treatment with CspB.31,32 Cells in the logarithmic growth phase were seeded in 6-well plates at a density of 1 × 105 cells/mL and treated with varying concentrations of CspB (32, 64, and 100 μM for PC3 and DU145 cells, and 50, 100, and 150 μM for LNCaP cells). Following treatment, the cells were harvested and fixed in 70% ice-cold ethanol overnight. The cells were washed twice with precooled PBS, centrifuged, and resuspended in a solution containing 100 μg/mL RNase A and 0.2% Triton X-100 for 30 min at 37 °C in a water bath. Subsequently, 400 μL of a 50 μg/mL propidium iodide (PI) staining solution was added, and the mixture was vortexed to ensure thorough mixing. The cells were incubated at room temperature in the dark for 30 min. Flow cytometry was utilized to analyze the DNA content, and the distribution of cells across the various phases of the cell cycle was evaluated using FlowJo software (Becton, Dickinson & Company). During the analysis, cells that adhered to the cell cycle were excluded. Approximately 10 000 cells were analyzed for each sample and processed at a low flow rate. The results were derived from three samples across three independent experiments.
Quantitative real-time PCR (qPCR) analysis
Following treatment with varying concentrations of CspB, the cells were harvested using trypsin, and total RNA was extracted from three distinct cell lines utilizing the Animal Tissue/Cell Total RNA Extraction Kit (Zhuangmeng, Beijing, China). The purity and concentration of the extracted RNA were assessed using a NanoDrop One spectrophotometer (Thermo Fisher Scientific). RNA purity was evaluated based on the absorbance ratios of A260/280 and A260/230. A ratio of A260/280 between 1.8 and 2.0 and an A260/230 ratio between 2.0 and 2.2 indicated high-quality RNA. For the first-strand reverse transcription, 2 μg of high-quality RNA was converted into cDNA using a high-temperature-resistant reverse transcriptase kit (M-MLV). Subsequently, agarose gel electrophoresis was performed to validate the quality of the resulting products. Quantitative polymerase chain reaction (qPCR) analysis of the three cell lines was conducted using the 2 × HQ SYBR qPCR Mix (High ROX) reagent. The primer sequences for the target genes and the optimal experimental conditions are detailed in Table 1. Gene expression levels were quantified using the 2-ΔΔCt method, with GAPDH as an internal reference.33,34
Table 1.
Primer sequence for qPCR
|
Primer
|
Primer sequence (5'-3')
|
PCR product size(bp)
|
TM (°C)
|
| DP1-F |
AGGGCCTACGGCATTTCTC |
83 |
59.47 |
| DP1-R |
CTCGTCTGCCACTTCGTTGT |
60.6 |
| CDKN2C-F |
GGGGACCTAGAGCAACTTACT |
81 |
58.53 |
| CDKN2C-R |
CAGCGCAGTCCTTCCAAAT |
58.44 |
| CDK6-F |
TCTTCATTCACACCGAGTAGTGC |
130 |
60.61 |
| CDK6-R |
TGAGGTTAGAGCCATCTGGAAA |
58.82 |
| E2F1-F |
CATCCCAGGAGGTCACTTCTG |
145 |
59.79 |
| E2F1-R |
GACAACAGCGGTTCTTGCTC |
59.76 |
| CDKN1A-F |
TGTCCGTCAGAACCCATGC |
139 |
60.0 |
| CDKN1A-R |
AAAGTCGAAGTTCCATCGCTC |
58.66 |
| GAPDH-F |
ACAACTTTGGTATCGTGGAAGG |
101 |
58.59 |
| GAPDH-R |
GCCATCACGCCACAGTTTC |
59.79 |
Western blotting
The expression of cell cycle-associated marker proteins in various cell lines treated with CspB was evaluated using Western blot analysis. In brief, after treatment with different concentrations of CspB, 200 µL of RIPA lysis buffer (comprising 50 mM Tris [pH 7.6], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS) supplemented with a mixture of phosphatase and protease inhibitors was added to each well of a 6-well plate. Following thorough lysis and centrifugation at 10,000 g for 10 min, the resulting supernatant was collected for subsequent experimental analyses. Protein quantification was performed using a BCA kit. Subsequently, 25 µg of each sample was loaded onto a 12% SDS polyacrylamide gel and subjected to electrophoresis. After separation on the gel, the proteins were electro-transferred to a nitrocellulose membrane at 100 V for 40 min. The membrane was then blocked with 5% non-fat powdered milk for 1 hour at room temperature and subsequently incubated overnight at 4 °C with the appropriate primary antibodies.35,36 The primary antibodies utilized in this study included those against CDK6 (cat. no. ET1612-3; 1:500; HUABIO), E2F1 (cat. no. ET1701-73; 1:500; HUABIO), CDKN2C (cat. no. ER64492; 1:1,000; HUABIO), DP1 (cat. no. ET7110-43; 1:500; HUABIO) and β-actin (cat. no. AF7018; 1:3,000; Affinity Biosciences). The PVDF membranes (Millipore) were washed five times with TBST (3 min each time). Subsequently, the membranes were incubated at room temperature for 1 hour with a 1:1,000 dilution of horseradish peroxidase-conjugated anti-rabbit secondary antibody (cat. no. A0208; 1:1,000; Beyotime Institute of Biotechnology). Following this, the membranes underwent an additional round of washing five times with TBST (3 min each time). Finally, the membranes were co-incubated with a chemically enhanced luminescence substrate for 1 min and chemiluminescence (Affinity Biosciences) was detected by exposing the membranes to a gel imager (Tanon, China). The grey values of the Western blot protein bands were analyzed using Image J (National Institutes of Health).
Statistical analysis
The experiments were conducted in triplicate, and the results are presented as the mean ± standard deviation (SD). Statistical analysis of mean differences was performed using a one-way analysis of variance, followed by a Bonferroni post hoc test, utilizing GraphPad Prism software version 8.0 (San Diego, CA, USA). P < 0.05 was considered to indicate a statistically significant difference.
Results
NMR analysis of CspB
About 1.1 g of monomeric compounds were obtained by serial separation purification and preparative liquid chromatography preparation. The NMR hydrogen and carbon spectrum show that the compound has the typical characteristics of flavonoids (Figs. S1 and S2). It was found that the NMR data were consistent with the data of CspB from the literature,37 and through HPLC verification (Fig. 1a), the compound was identified as CspB; the chemical structure is shown in Fig. 1b. The hydrogen spectral data of the compound were: δH 12.68 (1H, brs, 5-OH), 9.97 (1H, brs, 4'-OH), 7.69 (1H, s, H-2′), 7.65 (1H, d, J = 8.0Hz, H-6′), 6.97(1H, d, J = 8.0Hz, H-5), 6.93(1H, s, H-8), 3.93 (3H, s, OCH3), 3.88 (3H, s, OCH3), 3.82(3H, s, OCH3), 3.74 (3H, s, OCH3); and the carbon spectrum data were: δC 178.7 (C-4), 159.1 (C-7), 156.2 (C-9), 152.2 (C-2), 152.1 (C-5), 150.3 (C-3'), 147.9 (C-4'), 138.2 (C-3), 132.0 (C-6), 122.8(C-1'), 121.1 (C-6'), 116.1 (C-5'), 112.5 (C-2'), 106.0 (C-10), 91.9 (C-8), 60.5 (C-OCH3), 60.1 (C-OCH3), 56.9 (C-OCH3), 56.2 (C-OCH3).
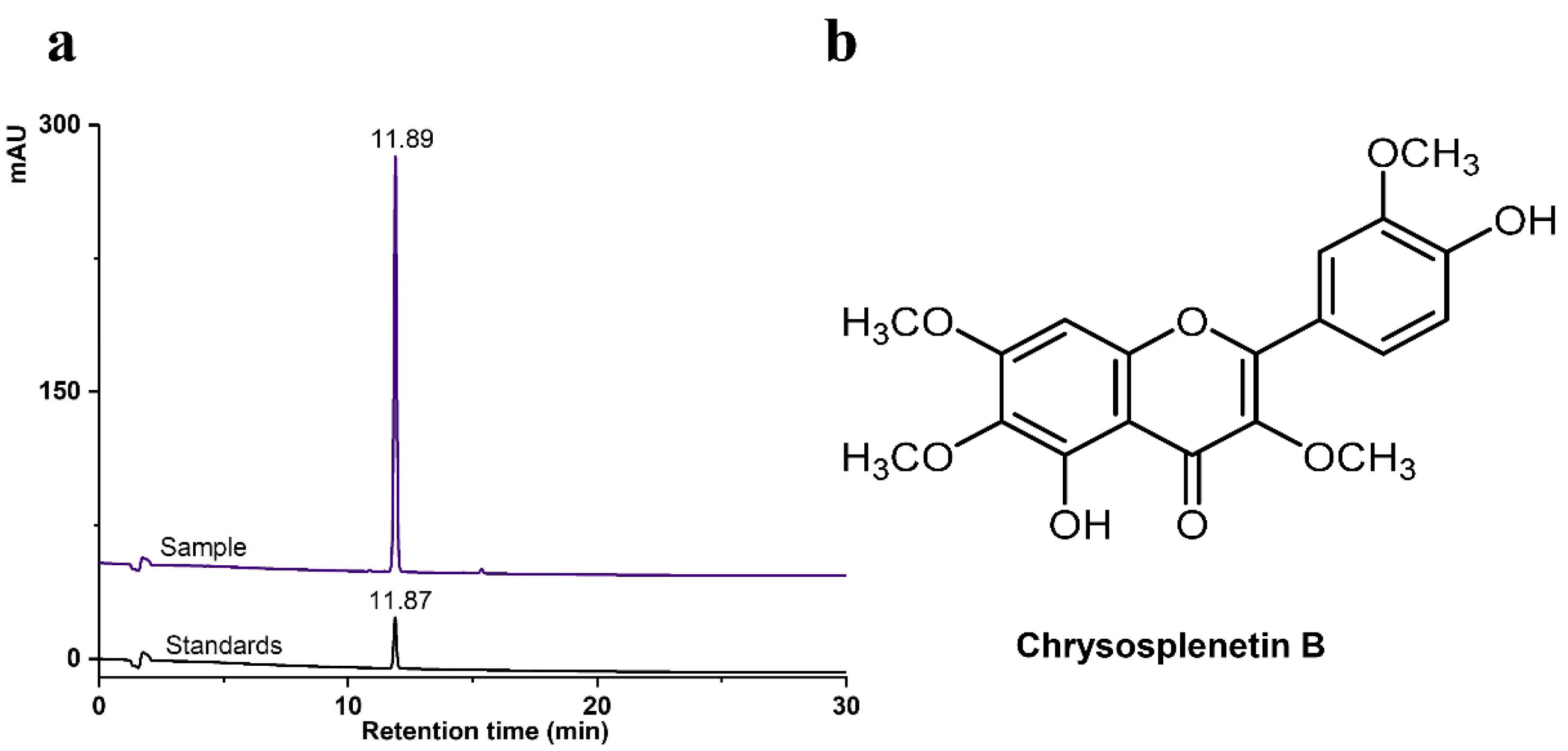
Fig. 1.
HPLC spectra of CspB. (a) HPLC chromatograms of CspB; (b) chemical structure of CspB.
.
HPLC spectra of CspB. (a) HPLC chromatograms of CspB; (b) chemical structure of CspB.
CspB inhibits the viability of PCa cells
Cell viability was assessed after treatment for 12, 24 and 48 h. CspB induced a dose-dependent reduction in cell viability in all PCa cell lines (Fig. 2). The IC50 values after treatment for 24 h are summarised in Table 2. Furthermore, it was observed that the inhibitory effect of CspB on PC3 and DU145 cells exhibited a significantly greater magnitude compared with its effect on LNCaP cells under identical treatment conditions. Our results suggested that advanced PCa cells, such as PC3 and DU145, exhibit higher sensitivity towards CspB. Therefore, the PC3 cell line was employed for subsequent investigations, including transcriptome analysis and exploration of cell death mechanisms at the transcriptomic level.
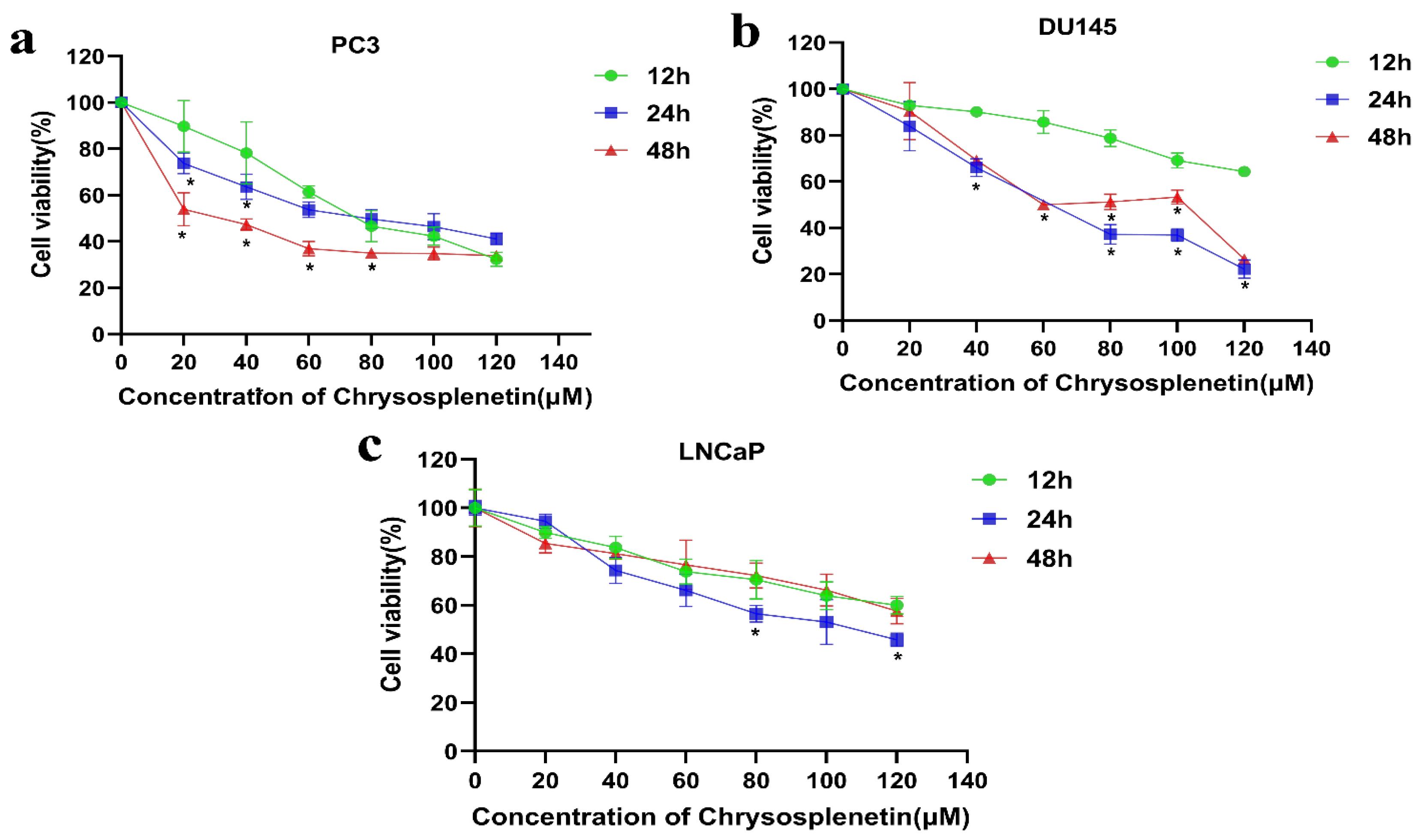
Fig. 2.
CspB inhibits prostate cancer cell viability in a dose-dependent manner. Panels (a-c) display the viability of PC3, DU145, and LNCaP cell lines across treatment groups. One-way ANOVA analysis revealed a significant difference from the 12-hour treatment group at *P < 0.05.
.
CspB inhibits prostate cancer cell viability in a dose-dependent manner. Panels (a-c) display the viability of PC3, DU145, and LNCaP cell lines across treatment groups. One-way ANOVA analysis revealed a significant difference from the 12-hour treatment group at *P < 0.05.
Table 2.
IC50 values of prostate cancer cells after 24h of CspB
|
Cell line
|
IC50 (95% CI)
|
R2
|
| PC3 |
64.69 µM |
0.9024 |
| DU145 |
73.45 µM |
0.9339 |
| LNCaP |
103.43 µM |
0.9247 |
RNA sequencing and pathway analysis of PCa cells treated with CspB
Transcriptome analysis detected approximately 14,560 genes in the samples. Volcano plots and heat maps were generated to visualize the gene expression patterns (Fig. 3a, 3b). Subsequently, we conducted a differential gene analysis to identify upregulated and downregulated genes in PC3 cells before and after CspB treatment. Our findings demonstrated that compared with the control, there were 2,570 differentially expressed genes (q-value < 0.05 and |log2FC| > 1) in PC3 cells following CspB treatment; 1 073 genes were upregulated while 1 497 genes were downregulated.
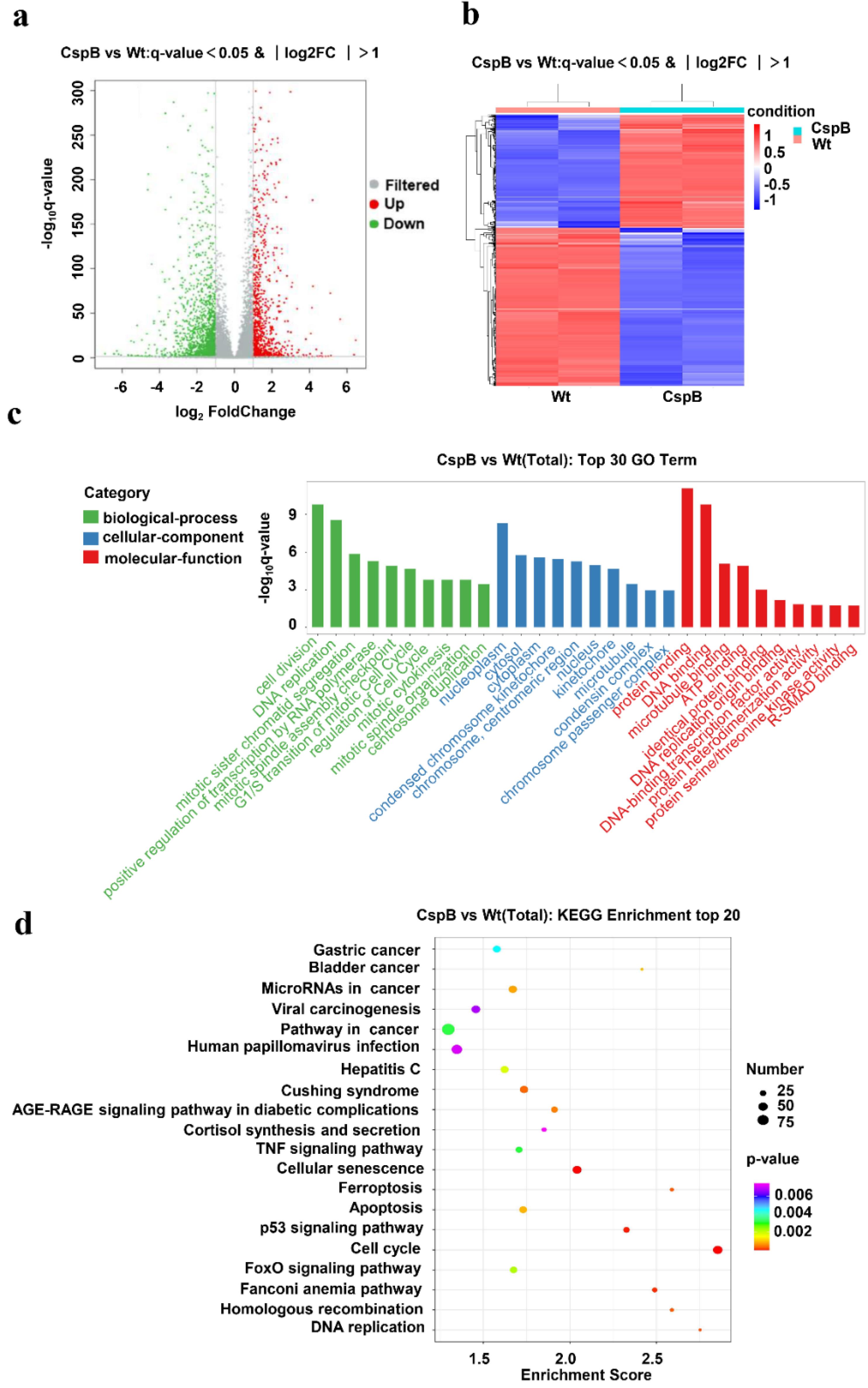
Fig. 3.
RNA sequencing and pathway analysis of prostate cancer cells treated with CspB. (a) A volcano plot illustrating the differentially expressed genes between the CspB-treated and wild-type (Wt) groups. (b) Cluster analysis of the differentially expressed genes in CspB-treated PC3 cells. (c) GO analysis of the dysregulated genes in CspB-treated cells, with green representing biological processes, blue indicating cellular components and red denoting molecular functions. (d) KEGG analysis of the differentially expressed genes.
.
RNA sequencing and pathway analysis of prostate cancer cells treated with CspB. (a) A volcano plot illustrating the differentially expressed genes between the CspB-treated and wild-type (Wt) groups. (b) Cluster analysis of the differentially expressed genes in CspB-treated PC3 cells. (c) GO analysis of the dysregulated genes in CspB-treated cells, with green representing biological processes, blue indicating cellular components and red denoting molecular functions. (d) KEGG analysis of the differentially expressed genes.
To investigate the potential biological functions and associated pathways of the differentially expressed genes, we performed a GO enrichment analysis. This analysis categorized the functions of these genes into three levels: biological process, cellular component, and molecular function. The top 30 GO entries exhibiting significant differences at each level were identified based on P values and are presented in ascending order (Fig. 3c). Notably, the differentially expressed genes were found to be enriched in critical biological processes, including cell division, DNA replication, and mitotic sister chromatid segregation; essential cellular components, such as nucleoplasm and cytosol; and essential molecular functions, including protein binding and DNA binding.
Pathway analysis of differentially expressed genes utilizing the KEGG pathway database revealed the top 20 pathways significantly associated with these genes (Fig. 3d). The size of each bubble in the figure represents the number of differentially expressed genes within each respective pathway. Notably, pathways that exhibited higher enrichment scores and a more significant number of differentially expressed genes included the cell cycle (hsa04110), the p53 signalling pathway (hsa04115), the Fanconi anemia pathway (hsa03460), and cellular senescence (hsa04218). Among these, the cell cycle pathway (hsa04110) demonstrated an exceptionally high enrichment score and was linked to a substantial number of differentially expressed genes, indicating its potential significance in mediating the antiproliferative effects of CspB on PC3 cells.
CspB induces cell cycle arrest at the G1 phase in PCa cells
The observed inhibitory effect of CspB on the proliferation of the three PCa cell lines indicates that CspB may exert its antiproliferative activity through the modulation of the cell cycle, as suggested by transcriptome analysis. PI staining was conducted on the three PCa cell lines following 24 hours of drug administration, and the cell cycle distribution of the cells was subsequently analyzed using flow cytometry.
Treatment with CspB resulted in a G1 cell cycle arrest in PC3, DU145, and LNCaP PCa cells, while DU145 cells also demonstrated a G2 cell cycle arrest. Flow cytometric analysis indicated that the proportion of cells in the G1 phase increased in PC3 cells after 24 hours of CspB treatment, exhibiting a dose-dependent effect that ranged from 52.68% (0 µM) to 74.59% (32.4 µM) and 75.61% (64.7 µM). Similarly, the percentage of DU145 cells in the G1 phase increased from 54.66% (0 µM) to 82.38% (32.4 µM) and 72.01% (64.7 µM). Furthermore, there was a corresponding dose-dependent increase in the proportion of cells in the G2 phase. The S-phase cell population of LNCaP cells was significantly diminished, whereas G1-phase cells exhibited a notable accumulation following treatment with varying doses of CspB. Specifically, the proportions of G1-phase cells were recorded as 72.63% (0 µM), 89.97% (50 µM), and 89.2% (100 µM), respectively (Fig. 4a, 4b). Cell proliferation and cell cycle progression are intricately interconnected, with cell cycle regulation serving as an effective mechanism for controlling cellular proliferation.38,39 Numerous anticancer agents have been shown to impede cancer cell cycle progression in the G1, S, or G2/M phases, leading to sub-G1 phase accumulation and subsequent apoptosis. These findings suggest that CspB may influence G1 cell cycle progression in PC3, DU145, and LNCaP cells, potentially contributing to its anticancer efficacy against prostate cancer cells.
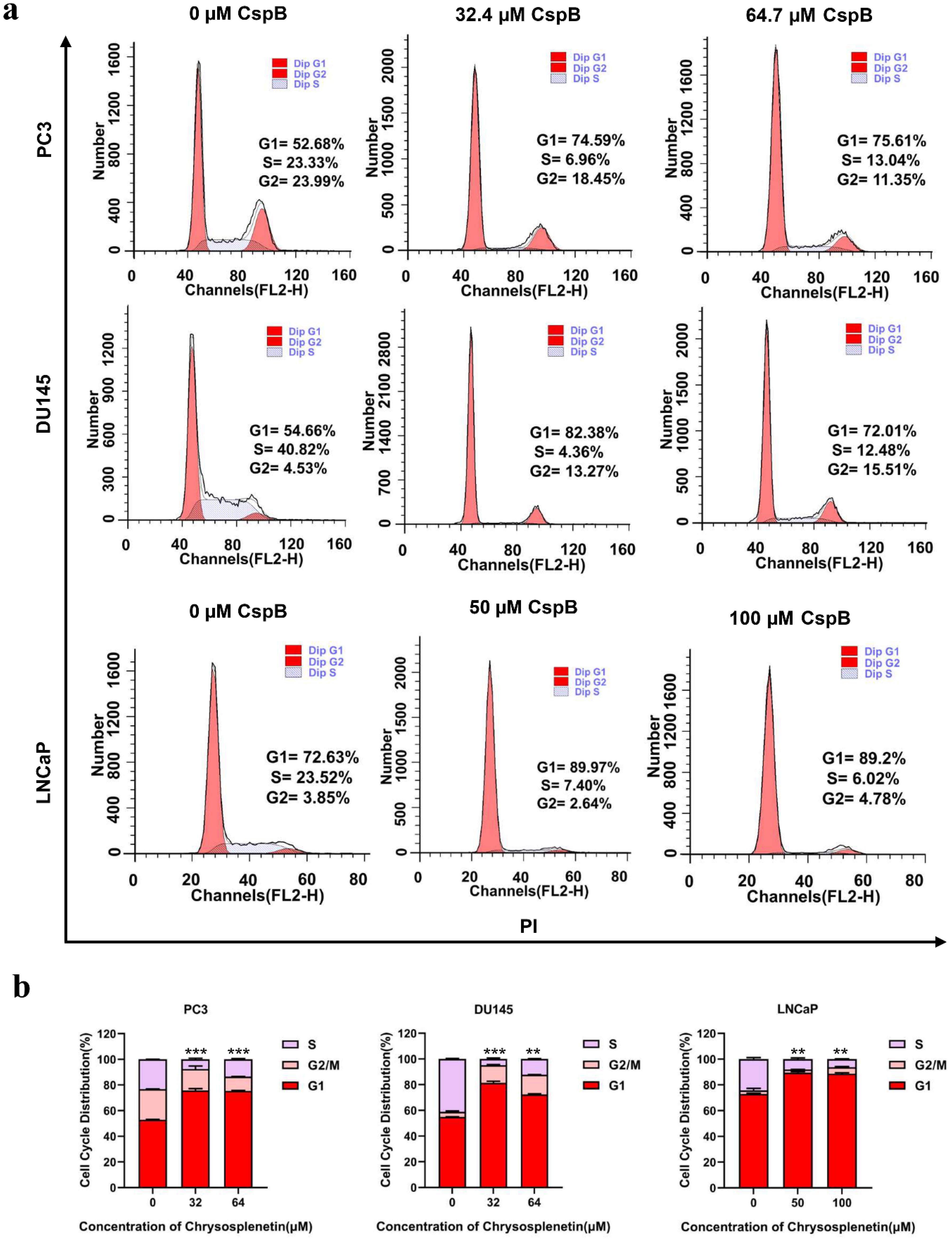
Fig. 4.
CspB induces cell cycle arrest at the G1 phase in prostate cancer cells. (a) Flow cytometric analysis of three prostate cancer cell lines undergoing G1 cycle arrest after 24 hours of CspB treatment. The histograms depict the distribution of cells in the G1 (left red segment), S (grey segment), and G2/M (right red segment) phases of the cell cycle. (b) Proportions of cells in each phase of the cell cycle for three prostate cancer cell lines following treatment with varying concentrations of CspB. Statistical significance was determined using one-way ANOVA (*P < 0.05, ** P <0.01, *** P < 0.001) compared to the control group.
.
CspB induces cell cycle arrest at the G1 phase in prostate cancer cells. (a) Flow cytometric analysis of three prostate cancer cell lines undergoing G1 cycle arrest after 24 hours of CspB treatment. The histograms depict the distribution of cells in the G1 (left red segment), S (grey segment), and G2/M (right red segment) phases of the cell cycle. (b) Proportions of cells in each phase of the cell cycle for three prostate cancer cell lines following treatment with varying concentrations of CspB. Statistical significance was determined using one-way ANOVA (*P < 0.05, ** P <0.01, *** P < 0.001) compared to the control group.
Effects of CspB on G1 phase-related gene expression
The results of the flow cytometry experiments indicated that CspB induced cell cycle arrest at the G1 phase in PC3, DU145, and LNCaP prostate cancer cells, demonstrating inhibitory effects on cancer cell proliferation. qPCR was subsequently utilized to assess the expression levels of genes associated with the G1 cell cycle phase. Total mRNA was isolated from PC3, DU145, and LNCaP cells treated with varying concentrations of CspB, followed by an analysis of the relative expression levels of cell cycle-related genes, including DP1 (TFDP1), CIP1 (CDKN1A/P21), KIP1 (CDKN1B/P27), and members of the INK4 family: CDKN2B (P15INK4B), CDKN2C (P18INK4C), CDKN2D (P19INK4D), CDK6, and E2F1. As illustrated in Figs. 5-7, the relative mRNA levels of DP1, CDKN2C, CDK6, and E2F1 in androgen-independent PCa cells (PC3 and DU145) were significantly reduced in a dose-dependent manner following 24 hours of treatment with CspB (P < 0.001), compared to the control group. Furthermore, there was a significant dose-dependent increase in the mRNA levels of KIP1 (P27) and CIP1 (P21) (P < 0.05). The expression patterns observed in CspB-treated androgen-dependent PCa cells (LNCaP) were consistent with those in PC3 and DU145 cells. The concordance between qPCR and transcriptome sequencing data suggests a potential relationship between the antiproliferative effects of CspB on PCa cells and G1 phase cell cycle arrest.
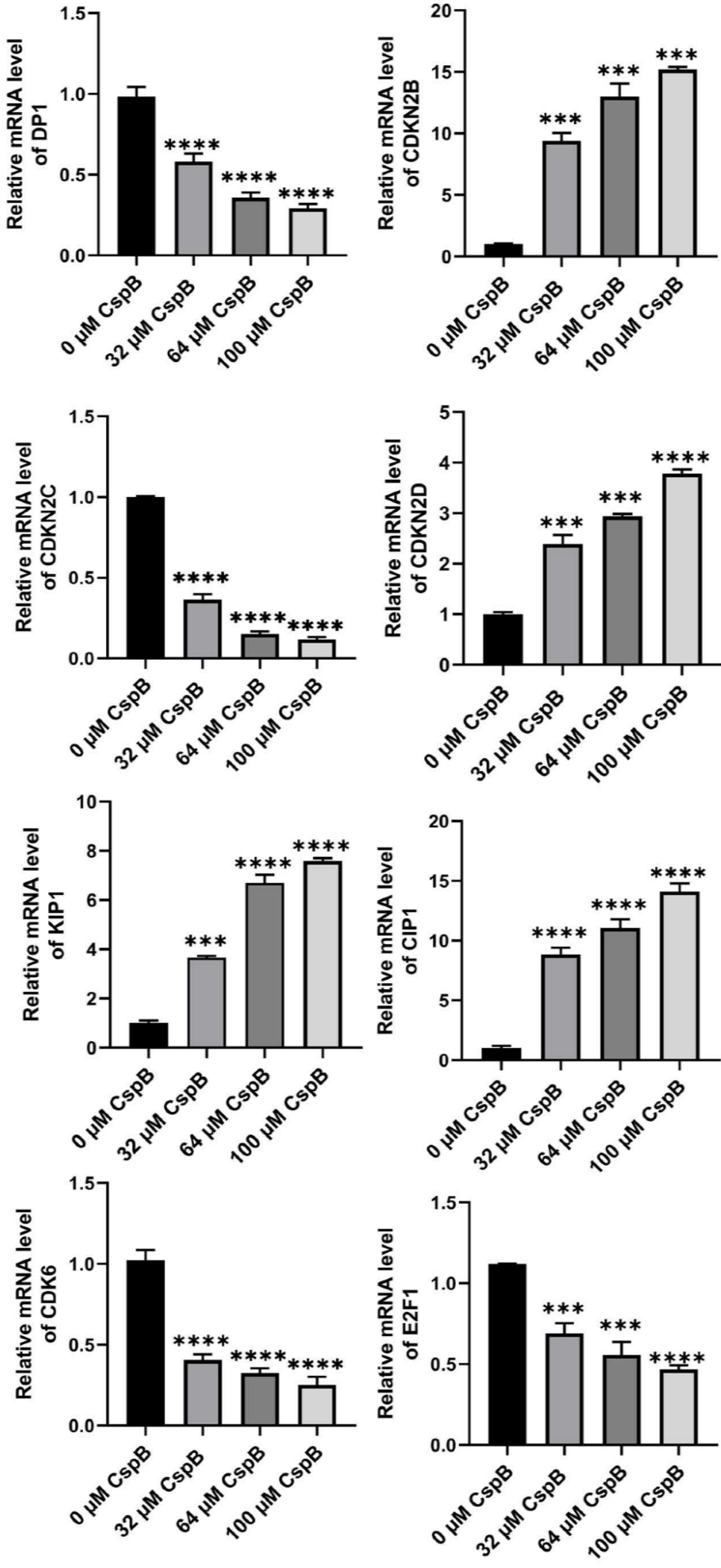
Fig. 5.
Effects of CspB on the expression of G1 phase-related genes in PC3 cells was investigated. The relative mRNA expression levels in prostate cancer cells were assessed using qRT-PCR. The data are presented as the mean ± SD (n = 4). Statistical analysis was conducted utilizing one-way analysis of variance (ANOVA). * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001.
.
Effects of CspB on the expression of G1 phase-related genes in PC3 cells was investigated. The relative mRNA expression levels in prostate cancer cells were assessed using qRT-PCR. The data are presented as the mean ± SD (n = 4). Statistical analysis was conducted utilizing one-way analysis of variance (ANOVA). * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001.
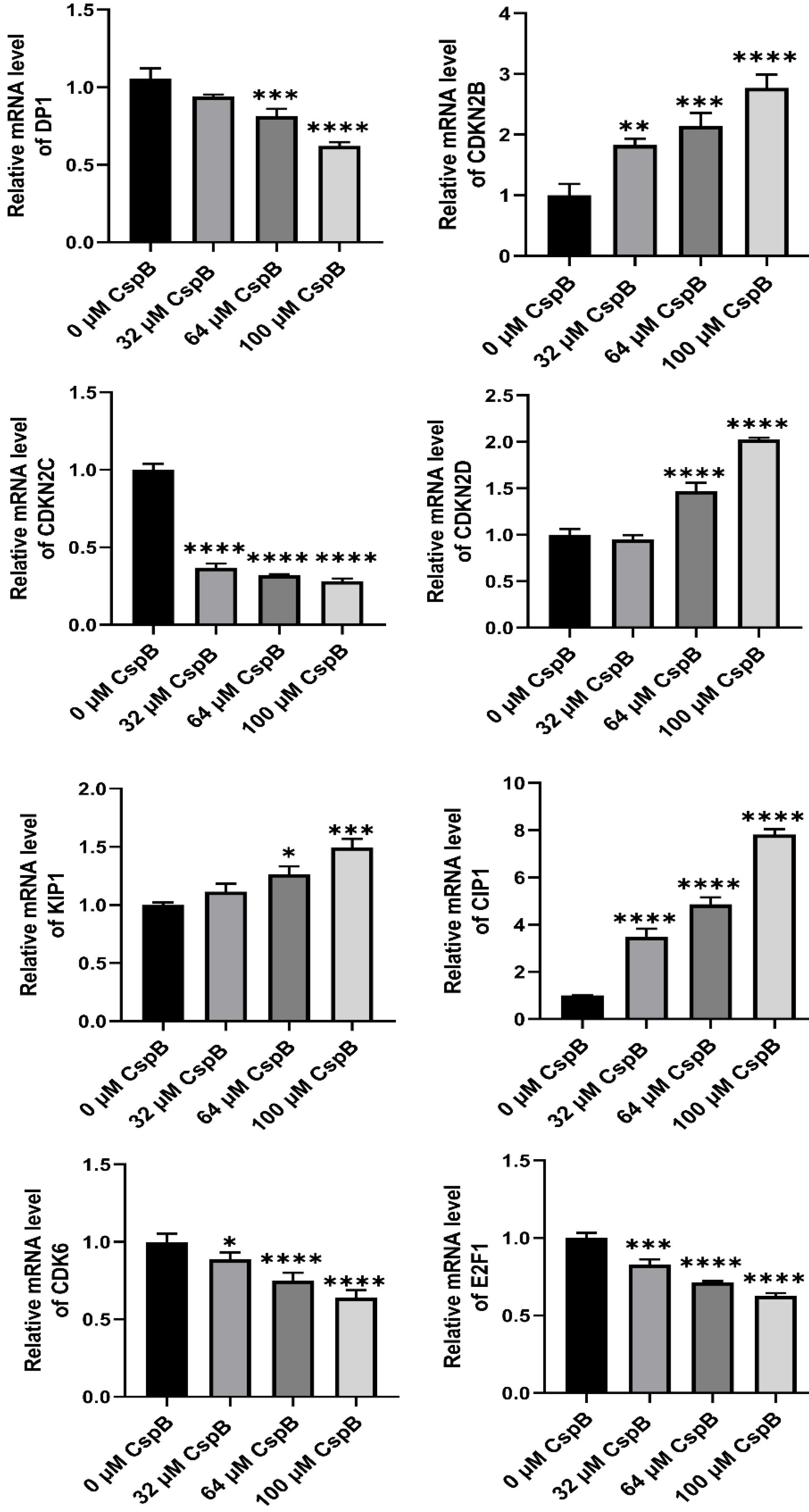
Fig. 6.
Effects of CspB on G1 phase-related gene expression in DU145 cells. The relative mRNA expression in prostate cancer cells was analyzed using qRT-PCR. The control group was used for comparison. Data are shown as the mean ± SD (n = 4). Statistical analysis was performed using one-way ANOVA. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001.
.
Effects of CspB on G1 phase-related gene expression in DU145 cells. The relative mRNA expression in prostate cancer cells was analyzed using qRT-PCR. The control group was used for comparison. Data are shown as the mean ± SD (n = 4). Statistical analysis was performed using one-way ANOVA. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001.
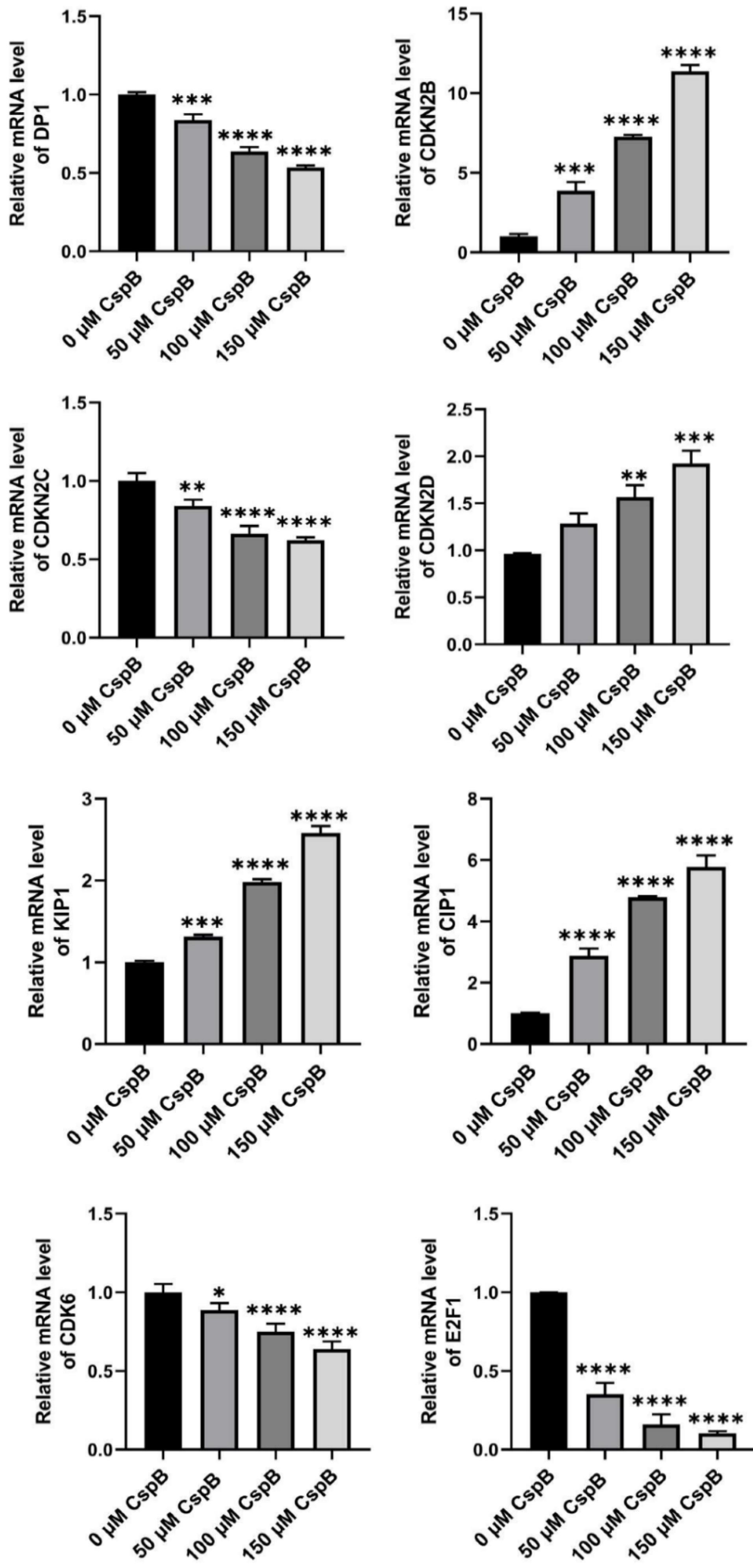
Fig. 7.
Effects of CspB on G1 phase-related gene expression in LNCap cells. The relative mRNA expression in prostate cancer cells was analyzed using qRT-PCR. The control group was used for comparison. Data are shown as the mean ± SD (n = 4). Statistical analysis was performed using one-way ANOVA. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001.
.
Effects of CspB on G1 phase-related gene expression in LNCap cells. The relative mRNA expression in prostate cancer cells was analyzed using qRT-PCR. The control group was used for comparison. Data are shown as the mean ± SD (n = 4). Statistical analysis was performed using one-way ANOVA. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001.
Effects of CspB on G1 phase-related protein expression
Western blotting was employed to assess the expression of essential proteins involved in the regulation of the G1 phase of the cell cycle, with the results presented in Fig. 8. In comparison to the control group, treatment with CspB led to a dose-dependent and statistically significant reduction in the protein expression levels of DP1 and CDKN2C (P18INK4C), with a more pronounced decrease noted at higher concentrations of CspB (P < 0.01). This effect was particularly evident in PC3 and DU145 cell lines. In PC3 cells, only the high-dose treatment resulted in a significant reduction in CDK6 protein expression levels when compared to the control group (P< 0.05). Conversely, both medium-dose and high-dose treatments significantly decreased CDK6 protein expression levels in LNCaP and DU145 cells (P < 0.01). Additionally, CspB treatment significantly diminished E2F1 protein expression levels in both PC3 and LNCaP cells, with significant reductions also observed following medium-dose and high-dose treatments in DU145 cells (P < 0.001).
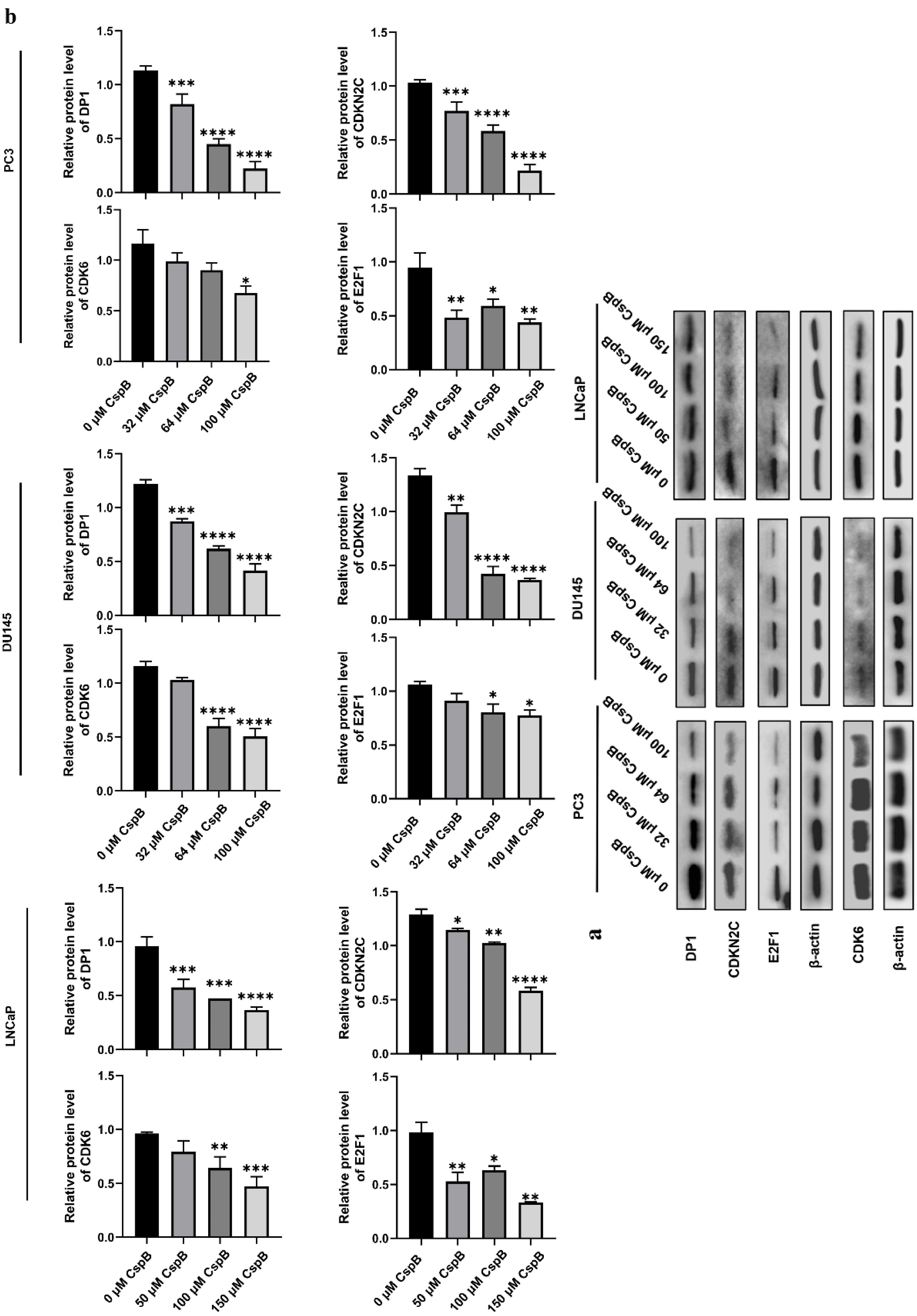
Fig. 8.
Effects of CspB on G1 phase-related protein expression. (a) Protein expression of DP1, CDKN2C, CDK6 and E2F1 in prostate cancer cells was detected by western blotting. (b) Data analysis of DP1, CDKN2C, CDK6 and E2F1 protein expression with β-actin as an internal reference.
.
Effects of CspB on G1 phase-related protein expression. (a) Protein expression of DP1, CDKN2C, CDK6 and E2F1 in prostate cancer cells was detected by western blotting. (b) Data analysis of DP1, CDKN2C, CDK6 and E2F1 protein expression with β-actin as an internal reference.
Discussion
PCa is one of the most prevalent malignancies, posing a significant clinical challenge due to reduced cellular androgen sensitivity. The diminished or absent response to first-line ADT severely limits the efficacy of available treatments.40 Tumour cell resistance poses a significant challenge to the efficacy of chemotherapeutic agents in cancer treatment, whereas traditional Chinese herbs and their bioactive constituents exhibit comparable effectiveness to chemotherapeutic drugs. In this study, we isolated and identified CspB, a flavonol compound, from the herb Laggera pteridonta, commonly used in Yunnan, China, intending to identify potent novel drugs for the treatment of CRPC. Cell viability experiments demonstrated that treatment with CspB effectively suppressed the growth of LNCap, DU145 and PC3 PCa cells with varying degrees of androgen dependence. After treatment with CspB, DU145 and PC3 cells exhibited increased sensitivity to the compound, as evidenced by lower IC50 values of 73.45 and 64.69 µM, respectively. However, the inhibitory activity of CspB was slightly diminished against DU145 cells, potentially due to the higher metastatic potential observed in PC3 cells. Conversely, LNCap cells displayed insensitivity to CspB treatment with an IC50 value of 103.43 µM. This discrepancy in sensitivity may be attributed to the fact that LNCap is an androgen-dependent PCa cell line (castration-sensitive), while both DU145 and PC3 are non-androgen-dependent PCa cell lines (castration-resistant). These variations align with similar differences observed in the inhibitory activities of other compounds. The response of PC3 and DU145 cells to curcumin derivatives RL118 and RL121 has been reported to vary, with PC3 cells exhibiting greater sensitivity.41 Aloperine has been shown to have a weaker inhibitory effect on DU145 and PC3 cells compared with LNCap cells by Ling et al.42 Sheng et al43 reported that LNCap cells had the lowest sensitivity to Curcumol among the three cell lines tested, including DU145 and PC3. Aggarwal et al demonstrated that PEITC had the most potent inhibitory effect on DU145 cell proliferation but the weakest on LNCap cells.44 Vakhrusheva et al revealed that Artesunate was most effective against PC3 cells, while LNCap and DU145 cells exhibited similar sensitivity levels.45 These findings indicate that there may be variations in response to drugs between PC3, DU145 and LNCaP cells, which can be attributed to the increased metastatic capability of PC3. Additionally, CspB demonstrates enhanced suppression of highly malignant tumour cells while exhibiting reduced toxicity towards normal cells, thus displaying promising potential for therapeutic drug development. Previous studies have revealed that CspB exerts a pronounced cytotoxic effect on HeLa, A549, MCF-7, and T47D tumour cells while demonstrating minimal toxicity towards EVC304, Vero, HUVECs, and other normal cell lines.21
Numerous studies have demonstrated that flavonoids in natural products possess the ability to enhance the expression of CDK interaction protein (CIP/P21)/kinase inhibitor protein (KIP/P27) and the INK4 family of CDK inhibitors, thereby inducing G1 cell cycle arrest in tumour cells.46-49 The KEGG Enrichment analysis reveals significant enrichment of genes associated with cell cycle events among the top 20 results for differentially expressed genes, suggesting that CspB primarily functions by closely interacting with the cell cycle to inhibit the growth of CRPC cells. Following a 24-hour treatment of CspB, there is a noticeable increase in the number of cells in the G1 phase compared to the control group. Additionally, both mRNA and protein levels of CDK6 and E2F1 exhibit significant downregulation and a significant dose-dependent increase in the mRNA levels of KIP1 (P27) and CIP1 (P21). Transcriptome and flow cytometric analyses revealed that CspB induced cell cycle arrest at the G1 phase. The collaboration between CDKs and cyclins is indispensable for precisely regulating the cell cycle through substrate phosphorylation, facilitation of distinct phases, and initiation of DNA replication. Specifically, CDK4/6 phosphorylates the retinoblastoma gene (RB), releasing E2F and subsequent transcriptional activation of genes associated with cell cycle progression. This process effectively promotes the transition from the G1 to S phase and facilitates DNA replication.50
The ability of RB to modulate E2F family transcription factor activity, which controls the production of proteins essential for cellular replication, is vital for preventing tumour development. Consistent with these findings, levels of E2F1, the most extensively studied member of the E2F family in PCa, increase during the transition from benign prostate to localized PCa and further escalate in metastatic lymph nodes from first-treatment patients as well as CRPC.51,52
p53, a pivotal transcription factor that orchestrates cell cycle arrest and apoptosis in response to DNA damage. However, CRPC prostate cancer cells exhibit either absence or mutation of p53 expression, rendering P53 regulation ineffective in achieving cell cycle arrest.53 The analysis of differential genes indicates that fewer genes are involved in cell apoptosis while more genes are involved in the p53 signalling pathway. Therefore, how the P53 signalling pathway regulates cell death remains to be further studied.
We measured the expression levels of cell cycle-related genes and proteins to investigate the impact of varying doses of CspB on DNA replication and cell cycle progression in PC3, DU-145 and LNCaP PCa cells. Transcriptome and qPCR analyses revealed a significant upregulation of P21 and P27 gene levels, as well as an upregulation of INK4 family genes (except CDKN2C), while CDK6 gene levels were significantly downregulated compared with those in the control group. This finding is consistent with previous reports indicating that psoralidin exerts an inhibitory effect on the growth of CRPC cells by inducing cell cycle arrest at the G0/G1 phase through suppressing Cyclin/CDK complex activity and promoting the expression of p21 and p27.54 Furthermore, western blot analysis demonstrated a notable decrease in the protein expression levels of CDKN2C from the INK4 family and CDK6 from the CDK family following treatment with CspB compared with those observed in the control group. The significant reduction of CDKN2C at the gene and protein levels needs to be elucidated in subsequent studies.
The TFDP1 gene encodes DP1, a chaperone that forms a heterodimer with the transcription factor E2F.55 The tumour suppressor pRB targets E2F and plays vital roles in cell proliferation, apoptosis, differentiation, DNA repair and other biological processes. E2F1 is crucial for G1/S transition and S-phase progression.56 Additionally, a reduction was observed in the abundance of the transcription factor E2F. The release of transcription factor E2F1 was reduced, and western blotting results showed that the protein expression levels of DP1 and E2F1 transcription factors were significantly downregulated. This regulatory mechanism was consistent in androgen-dependent PCa LNCaP cells and in androgen-independent PCa PC3 and DU145 cells. Transcriptome analysis revealed that the expression of the CDKN1C gene was significantly upregulated. The protein encoded by the CDKN1C gene can tightly bind and strongly inhibit multiple G1 cyclin/CDK complexes and is a negative regulator of cell proliferation.57 Our study demonstrated that CspB extracted from Laggera pterodonta induced cell cycle arrest in the G1 phase in PCa cells. These findings suggested that treatment with CspB downregulated critical regulators involved in the transition of PCa cells from the G1 phase to the S phase, inhibiting DNA replication and impeding cell cycle progression. Consequently, a reduced cell viability effect was observed. Therefore, cell cycle arrest is a meaningful way to treat CRPC. More in-depth studies need to explore the specific target of action of CspB in PCa.
Conclusion
This study identified a flavonoid compound, CspB, isolated and purified from the traditional Chinese medicine Laggera pterodonta. The in vitro cell viability assay and transcriptome analysis revealed that CspB effectively suppressed the cell viability of LNCap, DU145 and PC3 cells by impeding cell cycle progression. Moreover, CspB exhibited a more pronounced inhibitory effect on the viability of PC3 and DU145 than LNCaP cells. Furthermore, the present study elucidated a putative mechanism by which CspB regulates the cell viability of PCa cells. Specifically, CspB was found to upregulate the expression of P21 and P27 in PC3 cells and to induce an increase in the negative G1 phase regulator gene CDKN1C. The current investigation primarily assessed the anticancer efficacy of CspB in vitro; however, further investigations are warranted to explore its impact in vivo.
Research Highlights
What is the current knowledge?
√ Prostate cancer often progresses to CRPC, leading to higher mortality and increased drug resistance. Therefore, developing new therapeutic agents is essential.
√ Chrysosplenetin B showed increased cell cycle arrest and apoptosis in cancer cells, while selectively inhibiting proliferation in breast cancer cell lines with low toxicity to normal cells.
√ The inhibitory potential of CspB on prostate cancer proliferation, particularly against CRPC cells, remains unexplored in the literature.
What is new here?
√ In vitro experiments showed that CspB inhibits the proliferation of prostate cancer cells, particularly the aggressive PC3 cells.
√ Transcriptome analysis showed that differentially expressed genes mainly affect the cell cycle, followed by ferroptosis.
√ CspB inhibits CRPC cell proliferation by suppressing cell cycle protein/CDK activity, upregulating P21 and P27, and inducing G1 phase arrest.
Competing Interests
The authors declare that there is no conflict of interest.
Ethical Approval
None to be declared.
Supplementary files
Supplementary file 1 contains Figs. S1 and S2.
(pdf)
References
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021; 71:209-49. doi: 10.3322/caac.21660 [Crossref] [ Google Scholar]
- Fabiani R, Minelli L, Bertarelli G, Bacci S. A western dietary pattern increases prostate cancer risk: a systematic review and meta-analysis. Nutrients 2016; 8:626. doi: 10.3390/nu8100626 [Crossref] [ Google Scholar]
- Feng Q, He B. Androgen receptor signaling in the development of castration-resistant prostate cancer. Front Oncol 2019; 9:858. doi: 10.3389/fonc.2019.00858 [Crossref] [ Google Scholar]
- Pejčić T, Todorović Z, Đurašević S, Popović L. Mechanisms of prostate cancer cells survival and their therapeutic targeting. Int J Mol Sci 2023; 24:2939. doi: 10.3390/ijms24032939 [Crossref] [ Google Scholar]
- Ryan CJ, Smith MR, de Bono JS, Molina A, Logothetis CJ, de Souza P. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med 2013; 368:138-48. doi: 10.1056/NEJMoa1209096 [Crossref] [ Google Scholar]
- Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med 2012; 367:1187-97. doi: 10.1056/NEJMoa1207506 [Crossref] [ Google Scholar]
- Smith MR, Saad F, Chowdhury S, Oudard S, Hadaschik BA, Graff JN. Apalutamide treatment and metastasis-free survival in prostate cancer. N Engl J Med 2018; 378:1408-18. doi: 10.1056/NEJMoa1715546 [Crossref] [ Google Scholar]
- Fujmoto S, Fujita K, Nishimoto M, Hamaguchi M, Kuwahara K, Hashimoto M. Sequential therapy with darolutamide in patients with non-metastatic castration-resistant prostate cancer resistant to enzalutamide or apalutamide. Cancer Med 2023; 12:3176-9. doi: 10.1002/cam4.5189 [Crossref] [ Google Scholar]
- He Y, Xu W, Xiao YT, Huang H, Gu D, Ren S. Targeting signaling pathways in prostate cancer: mechanisms and clinical trials. Signal Transduct Target Ther 2022; 7:198. doi: 10.1038/s41392-022-01042-7 [Crossref] [ Google Scholar]
- Li Y, Yang R, Henzler CM, Ho Y, Passow C, Auch B. Diverse AR gene rearrangements mediate resistance to androgen receptor inhibitors in metastatic prostate cancer. Clin Cancer Res 2020; 26:1965-76. doi: 10.1158/1078-0432.Ccr-19-3023 [Crossref] [ Google Scholar]
- Makarević J, Tsaur I, Juengel E, Borgmann H, Nelson K, Thomas C. Amygdalin delays cell cycle progression and blocks growth of prostate cancer cells in vitro. Life Sci 2016; 147:137-42. doi: 10.1016/j.lfs.2016.01.039 [Crossref] [ Google Scholar]
- Makarević J, Rutz J, Juengel E, Kaulfuss S, Reiter M, Tsaur I. Amygdalin blocks bladder cancer cell growth in vitro by diminishing cyclin A and cdk2. PLoS One 2014; 9:e105590. doi: 10.1371/journal.pone.0105590 [Crossref] [ Google Scholar]
- Zhang J. Research progress of Laggera pterodonta (DC) Benth. Pharmacy Information 2022; 11:359-71. doi: 10.12677/pi.2022.115047 [Crossref] [ Google Scholar]
- Xie YQ, Du XY, Liu D, Chen XQ, Li RT, Zhang ZJ. Chemical constituents from Laggera pterodonta and their anti-inflammatory activities in vitro. Phytochem Lett 2021; 43:126-9. doi: 10.1016/j.phytol.2021.04.001 [Crossref] [ Google Scholar]
- Patel DK. Biological potential and therapeutic benefit of chrysosplenetin: an applications of polymethoxylated flavonoid in medicine from natural sources. Pharmacol Res Mod Chin Med 2022; 4:100155. doi: 10.1016/j.prmcm.2022.100155 [Crossref] [ Google Scholar]
- Lan JE, Li XJ, Zhu XF, Sun ZL, He JM, Zloh M. Flavonoids from Artemisia rupestris and their synergistic antibacterial effects on drug-resistant Staphylococcus aureus. Nat Prod Res 2021; 35:1881-6. doi: 10.1080/14786419.2019.1639182 [Crossref] [ Google Scholar]
- Delnavazi MR, Saiyarsarai P, Jafari-Nodooshan S, Khanavi M, Tavakoli S, Hadavinia H. Cytotoxic flavonoids from the aerial parts of Stachys lavandulifolia Vahl. Pharm Sci 2018; 24:332-9. doi: 10.15171/ps.2018.47 [Crossref] [ Google Scholar]
- Yu X, Zhang Q, Tian L, Guo Z, Liu C, Chen J. Germacrane-type sesquiterpenoids with antiproliferative activities from Eupatorium chinense. J Nat Prod 2018; 81:85-91. doi: 10.1021/acs.jnatprod.7b00693 [Crossref] [ Google Scholar]
- Batra P, Sharma AK. Anti-cancer potential of flavonoids: recent trends and future perspectives. 3 Biotech 2013; 3:439-59. doi: 10.1007/s13205-013-0117-5 [Crossref] [ Google Scholar]
- Hong G, He X, Shen Y, Chen X, Yang F, Yang P. Chrysosplenetin promotes osteoblastogenesis of bone marrow stromal cells via Wnt/β-catenin pathway and enhances osteogenesis in estrogen deficiency-induced bone loss. Stem Cell Res Ther 2019; 10:277. doi: 10.1186/s13287-019-1375-x [Crossref] [ Google Scholar]
- Sinha S, Amin H, Nayak D, Bhatnagar M, Kacker P, Chakraborty S. Assessment of microtubule depolymerization property of flavonoids isolated from Tanacetum gracile in breast cancer cells by biochemical and molecular docking approach. Chem Biol Interact 2015; 239:1-11. doi: 10.1016/j.cbi.2015.06.034 [Crossref] [ Google Scholar]
- Liu N, Du P, Xiao X, Liu Y, Peng Y, Yang C. Microfluidic-based mechanical phenotyping of androgen-sensitive and non-sensitive prostate cancer cells lines. Micromachines (Basel) 2019; 10:602. doi: 10.3390/mi10090602 [Crossref] [ Google Scholar]
- Zhao Y, Cao J, Zhao J, Wei P, Wu R, Zhang J. Chemical analysis of Chrysosplenium from different species by UPLC-Q exactive orbitrap HRMS and HPLC-DAD. J Pharm Biomed Anal 2022; 218:114861. doi: 10.1016/j.jpba.2022.114861 [Crossref] [ Google Scholar]
- Huang SH, Tseng JC, Lin CY, Kuo YY, Wang BJ, Kao YH. Rooibos suppresses proliferation of castration-resistant prostate cancer cells via inhibition of Akt signaling. Phytomedicine 2019; 64:153068. doi: 10.1016/j.phymed.2019.153068 [Crossref] [ Google Scholar]
- Naito R, Kano H, Shimada T, Makino T, Kadomoto S, Iwamoto H. A new flavonoid derivative exerts antitumor effects against androgen-sensitive to cabazitaxel-resistant prostate cancer cells. Prostate 2021; 81:295-306. doi: 10.1002/pros.24106 [Crossref] [ Google Scholar]
- Yang X, Zhong Y, Wang D, Lu Z. A simple colorimetric method for viable bacteria detection based on cell counting Kit-8. Anal Methods 2021; 13:5211-5. doi: 10.1039/d1ay01624e [Crossref] [ Google Scholar]
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015; 12:357-60. doi: 10.1038/nmeth.3317 [Crossref] [ Google Scholar]
- Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. Improving RNA-seq expression estimates by correcting for fragment bias. Genome Biol 2011; 12:R22. doi: 10.1186/gb-2011-12-3-r22 [Crossref] [ Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015; 31:166-9. doi: 10.1093/bioinformatics/btu638 [Crossref] [ Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15:550. doi: 10.1186/s13059-014-0550-8 [Crossref] [ Google Scholar]
- Matsuoka T, Sugiyama A, Miyawaki Y, Hidaka Y, Okuno Y, Sakai H. Newly developed preclinical models reveal broad-spectrum CDK inhibitors as potent drugs for CRPC exhibiting primary resistance to enzalutamide. Cancer Sci 2024; 115:283-97. doi: 10.1111/cas.15984 [Crossref] [ Google Scholar]
- Dou B, Cui Y, Zhou Q, Fu J, Zhou Y, Zhang X. Mechanism of baicalein in treatment of castration-resistant prostate cancer based on network pharmacology and cell experiments. Front Pharmacol 2024; 15:1397703. doi: 10.3389/fphar.2024.1397703 [Crossref] [ Google Scholar]
- Qu M, Zhang G, Qu H, Vu A, Wu R, Tsukamoto H. Circadian regulator BMAL1::CLOCK promotes cell proliferation in hepatocellular carcinoma by controlling apoptosis and cell cycle. Proc Natl Acad Sci U S A 2023; 120:e2214829120. doi: 10.1073/pnas.2214829120 [Crossref] [ Google Scholar]
-
Cullot G, Boutin J, Fayet S, Prat F, Rosier J, Cappellen D, et al. Cell cycle arrest and p53 prevent ON-target megabase-scale rearrangements induced by CRISPR-Cas9. Nat Commun 2023. 14: 4072. doi: 10.1038/s41467-023-39632-w.
- Subhawa S, Naiki-Ito A, Kato H, Naiki T, Komura M, Nagano-Matsuo A. Suppressive effect and molecular mechanism of Houttuynia cordata Thunb extract against prostate carcinogenesis and castration-resistant prostate cancer. Cancers (Basel) 2021; 13:3403. doi: 10.3390/cancers13143403 [Crossref] [ Google Scholar]
- Zhang N, Wu W, Huang Y, An L, He Z, Chang Z. Citrus flavone tangeretin inhibits CRPC cell proliferation by regulating Cx26, AKT, and AR signaling. Evid Based Complement Alternat Med 2022; 2022:6422500. doi: 10.1155/2022/6422500 [Crossref] [ Google Scholar]
- Chougouo RD, Nguekeu YM, Dzoyem JP, Awouafack MD, Kouamouo J, Tane P. Anti-inflammatory and acetylcholinesterase activity of extract, fractions and five compounds isolated from the leaves and twigs of Artemisia annua growing in Cameroon. Springerplus 2016; 5:1525. doi: 10.1186/s40064-016-3199-9 [Crossref] [ Google Scholar]
- Foster DA, Yellen P, Xu L, Saqcena M. Regulation of G1 cell cycle progression: distinguishing the restriction point from a nutrient-sensing cell growth checkpoint(s). Genes Cancer 2010; 1:1124-31. doi: 10.1177/1947601910392989 [Crossref] [ Google Scholar]
- Ruijtenberg S, van den Heuvel S. Coordinating cell proliferation and differentiation: antagonism between cell cycle regulators and cell type-specific gene expression. Cell Cycle 2016; 15:196-212. doi: 10.1080/15384101.2015.1120925 [Crossref] [ Google Scholar]
- Ishii K, Sasaki T, Iguchi K, Kato M, Kanda H, Hirokawa Y. Pirfenidone, an anti-fibrotic drug, suppresses the growth of human prostate cancer cells by inducing G1 cell cycle arrest. J Clin Med 2019; 8:44. doi: 10.3390/jcm8010044 [Crossref] [ Google Scholar]
- Chen S, Nimick M, Cridge AG, Hawkins BC, Rosengren RJ. Anticancer potential of novel curcumin analogs towards castrate-resistant prostate cancer. Int J Oncol 2018; 52:579-88. doi: 10.3892/ijo.2017.4207 [Crossref] [ Google Scholar]
- Ling Z, Guan H, You Z, Wang C, Hu L, Zhang L. Aloperine executes antitumor effects through the induction of apoptosis and cell cycle arrest in prostate cancer in vitro and in vivo. Onco Targets Ther 2018; 11:2735-43. doi: 10.2147/ott.S165262 [Crossref] [ Google Scholar]
- Sheng W, Xu W, Ding J, Li L, You X, Wu Y. Curcumol inhibits the malignant progression of prostate cancer and regulates the PDK1/AKT/mTOR pathway by targeting miR-9. Oncol Rep 2021; 46:246. doi: 10.3892/or.2021.8197 [Crossref] [ Google Scholar]
- Aggarwal M, Saxena R, Asif N, Sinclair E, Tan J, Cruz I. p53 mutant-type in human prostate cancer cells determines the sensitivity to phenethyl isothiocyanate induced growth inhibition. J Exp Clin Cancer Res 2019; 38:307. doi: 10.1186/s13046-019-1267-z [Crossref] [ Google Scholar]
- Vakhrusheva O, Erb HH, Bräunig V, Markowitsch SD, Schupp P, Baer PC. Artesunate inhibits the growth behavior of docetaxel-resistant prostate cancer cells. Front Oncol 2022; 12:789284. doi: 10.3389/fonc.2022.789284 [Crossref] [ Google Scholar]
- Herrera-Sotero MY, Cruz-Hernández CD, Oliart-Ros RM, Chávez-Servia JL, Guzmán-Gerónimo RI, González-Covarrubias V. Anthocyanins of blue corn and tortilla arrest cell cycle and induce apoptosis on breast and prostate cancer cells. Nutr Cancer 2020; 72:768-77. doi: 10.1080/01635581.2019.1654529 [Crossref] [ Google Scholar]
- Khan F, Pandey P, Upadhyay TK, Jafri A, Jha NK, Mishra R. Anti-cancerous effect of rutin against HPV-C33A cervical cancer cells via G0/G1 cell cycle arrest and apoptotic induction. Endocr Metab Immune Disord Drug Targets 2020; 20:409-18. doi: 10.2174/1871530319666190806122257 [Crossref] [ Google Scholar]
- Lee H, Kim W, Kang HG, Kim WJ, Lee SC, Kim SJ. Geranium thunbergii extract-induced G1 phase cell cycle arrest and apoptosis in gastric cancer cells. Anim Cells Syst (Seoul) 2020; 24:26-33. doi: 10.1080/19768354.2019.1699161 [Crossref] [ Google Scholar]
- Zhou J, Li LU, Fang LI, Xie H, Yao W, Zhou X. Quercetin reduces cyclin D1 activity and induces G1 phase arrest in HepG2 cells. Oncol Lett 2016; 12:516-22. doi: 10.3892/ol.2016.4639 [Crossref] [ Google Scholar]
- Otto T, Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer 2017; 17:93-115. doi: 10.1038/nrc.2016.138 [Crossref] [ Google Scholar]
- Handle F, Prekovic S, Helsen C, Van den Broeck T, Smeets E, Moris L. Drivers of AR indifferent anti-androgen resistance in prostate cancer cells. Sci Rep 2019; 9:13786. doi: 10.1038/s41598-019-50220-1 [Crossref] [ Google Scholar]
- Ben-Salem S, Venkadakrishnan VB, Heemers HV. Novel insights in cell cycle dysregulation during prostate cancer progression. Endocr Relat Cancer 2021; 28:R141-55. doi: 10.1530/erc-20-0517 [Crossref] [ Google Scholar]
- Zhang Y, Xu L, Chang Y, Li Y, Butler W, Jin E. Therapeutic potential of ReACp53 targeting mutant p53 protein in CRPC. Prostate Cancer Prostatic Dis 2020; 23:160-71. doi: 10.1038/s41391-019-0172-z [Crossref] [ Google Scholar]
- Gulappa T, Reddy RS, Suman S, Nyakeriga AM, Damodaran C. Molecular interplay between cdk4 and p21 dictates G0/G1 cell cycle arrest in prostate cancer cells. Cancer Lett 2013; 337:177-83. doi: 10.1016/j.canlet.2013.05.014 [Crossref] [ Google Scholar]
- Nakajima R, Deguchi R, Komori H, Zhao L, Zhou Y, Shirasawa M. The TFDP1 gene coding for DP1, the heterodimeric partner of the transcription factor E2F, is a target of deregulated E2F. Biochem Biophys Res Commun 2023; 663:154-62. doi: 10.1016/j.bbrc.2023.04.092 [Crossref] [ Google Scholar]
- Saad D, Paissoni C, Chaves-Sanjuan A, Nardini M, Mantovani R, Gnesutta N. High conformational flexibility of the E2F1/DP1/DNA complex. J Mol Biol 2021; 433:167119. doi: 10.1016/j.jmb.2021.167119 [Crossref] [ Google Scholar]
- Stampone E, Caldarelli I, Zullo A, Bencivenga D, Mancini FP, Della Ragione F. Genetic and epigenetic control of CDKN1C expression: importance in cell commitment and differentiation, tissue homeostasis and human diseases. Int J Mol Sci 2018; 19:1055. doi: 10.3390/ijms19041055 [Crossref] [ Google Scholar]