Bioimpacts. 12(5):415-429.
doi: 10.34172/bi.2022.23336
Original Research
Silibinin exhibits anti-tumor effects in a breast cancer stem cell model by targeting stemness and induction of differentiation and apoptosis
Javad Firouzi 1, 2, 3  , Fattah Sotoodehnejadnematalahi 4, Alireza Shokouhifar 3
, Fattah Sotoodehnejadnematalahi 4, Alireza Shokouhifar 3  , Mahsa Rahimi 3, Niloufar Sodeifi 5, Parisa Sahranavardfar 3, Masoumeh Azimi 3, Ehsan Janzamin 3, Majid Safa 1, 2, 6, Marzieh Ebrahimi 3, *
, Mahsa Rahimi 3, Niloufar Sodeifi 5, Parisa Sahranavardfar 3, Masoumeh Azimi 3, Ehsan Janzamin 3, Majid Safa 1, 2, 6, Marzieh Ebrahimi 3, * 
Author information:
1Department of Tissue Engineering & Regenerative Medicine, Faculty of Advanced Technologies in Medicine, Iran University of Medical Sciences, Tehran, Iran
2Cellular and Molecular Research Centre, Iran University of Medical Sciences, Tehran, Iran
3Department of Stem Cells and Developmental Biology, Cell Science Research Center, Royan Institute for Stem Cell Biology and Technology, ACECR, Tehran 16635-148
4Department of Biology, School of Basic Science, Science and Research Branch, Islamic Azad University, 1477893855
5Department of Pathology, Reproductive Biomedicine Research Center, Royan Institute for Reproductive Biomedicine, ACECR, Tehran 16635-148, Iran
6Department of Hematology, Faculty of Allied Medicine, Iran University of Medical Sciences, Tehran, Iran
Abstract
Introduction:
Malignant breast cancer (BC) frequently contains a rare population of cells called cancer stem cells which underlie tumor relapse and metastasis, and targeting these cells may improve treatment options and outcomes for patients with BC. The aim of the present study was to determine the effect of silibinin on the self-renewal capacity, tumorgenicity, and metastatic potential of mammospheres.
Methods:
The effect of silibinin on viability and proliferation of MCF-7, MDA-MB-231 mammospheres, and MDA-MB-468 cell aggregation was determined after 72-120 hours of treatment. Colony and sphere formation ability, and the expression of stemness, differentiation, and epithelial-mesenchymal-transition (EMT)-associated genes were assessed by reverse transcription-quantitative polymerase chain reaction (qRT-PCR) in mammospheres treated with an IC50 dose of silibinin. Additionally, the antitumor capacity of silibinin was assessed in vivo, in mice.
Results:
The results of the present study showed that silibinin decreased the viability of all mammospheres derived from MCF-7, MDA-MB-231, and MDA-MB-468 cell aggregation in a dose-dependent manner. Colony and sphere-forming ability, as well as the expression of genes associated with EMT were reduced in mammospheres treated with silibinin. Additionally, the expression of genes associated with stemness and metastasis was also decreased and the expression of genes associated with differentiation were increased. Intra-tumoral injection of 2 mg/kg silibinin decreased tumor volumes in mice by 2.8 fold.
Conclusion:
The present study demonstrated that silibinin may have exerted its anti-tumor effects in BC by targeting the BC stem cells, reducing the tumorgenicity and metastasis. Therefore, silibinin may be a potential adjuvant for treatment of BC.
Keywords: Breast cancer stem cells, Silibinin, Mammospheres, Epithelial to mesenchymal transition
Copyright and License Information
© 2022 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
Breast cancer (BC) is the second leading cause of cancer-associated deaths in women worldwide,
1
and is often composed of a collection of different cell populations with varying properties.
2
Frequently, a small population of tumor cells with self-renewal capacity and resistance to commonly used therapeutics remain in patients who have been treated with conventional therapies, and these remaining cells are responsible for tumor growth, expansion, and metastasis.
3-5
Therefore, targeting and eliminating these tumor stem cells at the same time as the rest of the tumor may reduce relapse rates. However, developing a suitable model of cancer stem cells (CSCs) to be used in pharmacological studies remains a challenge. The majority of chemotherapeutics are initially tested two-dimensionally on a monolayer of tumor specific sorted stem cells based on their cell surface markers,
6
and this does not accurately reflect the tumor environment in patients. Another commonly used method for studying potential chemotherapeutics are xenograft mouse models or patient-derived xenograft models which are expensive and time-consuming, and numerous ethical concerns need to be taken into consideration, which may limit the scope of the studies.
7
Three dimensional (3D) tumor cell culture provides a more accurate representation of the tumor environment and is more likely to retain or replicate the proper signaling pathways, cell-cell interactions, and adhesions present in vivo.
8
Conclusions drawn from 3D tumor cultures may also be accurate as the exposure to drug treatments, stress, and oxygenation will be more representative of the tumor environment.
9
Additionally, development of tumor spheres in serum-free medium with reduced cell attachment properties may be useful in enriching CSCs. The aim of the present study was to use mammosphere models to investigate CSC richness, and to imitate tumor masses in vitro.
Over the past two decades, silymarin and its primary component silibinin, have been demonstrated to possess anticancer effects,
10
by targeting multiple events associated with tumor growth, including apoptosis,
11,12
cell cycle arrest,
12,13
inflammation,
14
angiogenesis,
15
cancer cell metabolism,
16,17
and invasion and metastasis,
18-20
with little to no toxic effects. Furthermore, silibinin combined with metformin enhanced the antiproliferative effects of metformin on BC cells through downregulation of cyclin D1 and hTERT gene expression,
21
and also enhanced the apoptotic effects of paclitaxel toxicity on various human gastric cancer cell lines.
22
Previous studies have demonstrated the potential of silibinin to target stemness and metastasis in bladder cancer through inhibition of the β-catenin/ZEB1 signaling pathway,
23
suppression of nuclear factor-κB activation in breast carcinoma,
24
by modulating interleukin 4/6-mediated survival signals in colon CSC-enriched spheroids.
25
Additionally, a combination of silibinin and spheroids reduced the phosphorylation of STAT3/ERK/AKT in CSCs of hepatocellular carcinoma.
26
Our previous study also showed the potential of silibinin to reduce stemness and induce apoptosis in 2D and 3D models of theMDA-MB-468 breast carcinoma cell line.
27
However, its effect on BC stem cells is unclear in vitro and in vivo. In the present study, mammospheres were used as a breast cancer stem cell (BCSC) model to determine the effect of silibinin on self-renewal capacity and the invasive potential. Furthermore, the effects of silibinin on tumor growth in vivo were evaluated. The results of the present study may improve our understanding of the mechanisms underlying the anti-cancer effects of silibinin in BC as well as improving our ability to specifically target the BC stem cells.
Materials and Methods
Cell lines and culture conditions
In the present study four BC cell lines were used: MCF-7 as a ER+, PR+ cell line; MDA-MB-231 cells, a poorly differentiated triple-negative BC (TNBC) cell line; MDA-MB-468 as a TNBC; and epithelial Mouse BC 4T1 cells, a 6-thioguanine resistant cell line (the passage number of all the cell lines cultures was three). All the human cell lines were purchased from the Iranian Biological Resource Center and the 4T1 cell line was purchased from Pasteur Institute of Iran. Human cell lines were cultivated in DMEM and 4T1 cells were cultured in RPMI-1640 supplemented with 2 mmol/L L-glutamine, 100 U/mL penicillin, 100 µg/mL streptomycin, non-essential amino acid, and 10% FBS (all purchased from Invitrogen; Thermo Fisher Scientific, Inc.). Cells were incubated with 5% CO2 at 37˚C. Trypsin/EDTA (Invitrogen; Thermo Fisher Scientific, Inc.) was used to harvest the cells.
Mammosphere culture
The mammospheres were formed using an anchorage-independent method. Flat-bottom 96-well plates and T25 flasks were coated with 12 mg/mL poly 2-hydroxyethyl methacrylate (Poly-HEMA; Sigma-Aldrich; Merck KGaA) in 95% ethanol and washed once with PBS before cell seeding. A total of 9 × 104 viable cells were transferred into a non-adherent T25 flask in 5 mL sphere medium including DMEM supplemented with basic fibroblast growth factor (bFGF; Royan Institute), 20 ng/mL epithelial growth factor (EGF; Royan Institute), and 2% B27 (Thermo Fisher Scientific, Inc), Fresh EGF, bFGF, and B27 were added every 2 days.
Cell viability and proliferation assay
The CellTiter 96 AQueous One Solution Cell Proliferation assay (Promega Corporation), which includes MTS was used according to the manufacturer’s protocol to evaluate cell viability. For assessment of cell viability, 1.5 × 103 cells/well were seeded into poly-HEMA-coated 96-well plates and cultivated in sphere medium for 96 hours to induce mammosphere formation. Subsequently, silibinin (Sigma-Aldrich; Merck KGaA) was dissolved in ethanol according to the manufacturer’s protocol, added to media to a final concentration of 0-1,600 µmol, and incubated for 72 hours at 37˚C with 5% CO2. For the cell proliferation assay, 2 × 103 mammosphere derived cells per well from the control (treated with ethanol alone) and the test group (treated with an IC50 dose of silibinin) were seeded in flat-bottom 96-well plates and incubated at 37˚C with 5% CO2 for 24, 48, 72, 96, and 120 hours. In both experiments, MTS was added to each well at the end of incubation time and further incubated for another 3 hours. Subsequently, 100 µL media from every well were transferred to a new flat-bottom 96-well plate and the optical density of the culture medium was measured at 490 nm using a microplate reader (Bio-Tek Instruments, Ltd.). Viability of cells in the mammospheres was calculated as a percentage as follows: (Average absorbance in the silibinin treated group/average absorbance in the control group) × 100. Viability assays were performed in triplicate.
Clonogenic assay
Mammospheres treated with an IC50 dose of silibinin for 72 hours were washed and dissociated using trypsin/EDTA. A total of 200 viable cells from both the control and treated groups were transferred into each well of a 6-well plate containing 2 mL supplemented DMEM. After 12 days of culturing, the medium was discarded, cells were washed with calcium and magnesium-free PBS, and fixed with 4% paraformaldehyde at 4˚C for 45 minutes. Colonies were stained with 200 µL 0.05% crystal violet for 5 minutes at room temperature (RT) and counted using an inverted light microscope at ×10 magnification. All results were normalized to the ethanol group. A 6-well plate was used for each replicate (two wells per a cell line) and 20-40 colonies were counted for each condition. At least three biological replicates were performed. For size analysis, all colonies in all nine experiments were analyzed using an inverted light microscope at ×10 magnification and cellSens Standard software version 1.5 (CellSens B.V.). Clonogenicity was calculated as follows: (Average number of colonies formed/number of cells seeded) × 100.
Mammosphere formation efficiency assay
Mammospheres treated with an IC50 dose of silibinin for 72 hours were washed and cells were dissociated using trypsin/EDTA. A total of 3 × 104 viable cells/well were plated in poly-HEMA-coated 6-well plates containing 2 mL sphere medium. Fresh EGF, bFGF, and B27 were added every 3 days. Mammospheres with diameters of 150-400 µm were counted after 7 days of culture. The mean diameter (d) of the mammospheres was determined using the following equation: d = (axb)1/2, where a and b are the orthogonal diameters of the spheroid.
28
Invasion and migration assay
Invasion and migration assays were performed in 24-well plates with 8.0 µm pore Transwell inserts (EMD Millipore). In order to investigate invasive ability, filters were pre-coated with 60 μL of diluted Matrigel (0.5 mg/mL; Sigma-Aldrich; Merck KGaA) for 12 hours. Uncoated filters were used for the migration assays. A total of 2.5×104 cells derived from treated (72 hours with an IC50 dose of silibinin) and control mammospheres in 200 µL serum-free culture medium was added to the filter inserts. Each filter was placed in the lower chamber with 600 µL culture medium containing 10% FBS. Filters were incubated at 37˚C with 5% CO2 for 12 hours. The cells which remained on the inside of the filter were removed using a cotton swab and cells attached to the bottom surface of filters were fixed with 4% paraformaldehyde at 4˚C for 45 minutes, and subsequently stained with a 0.5% crystal violet for 5 minutes at RT. The number of attached cells on the bottom surface of the filter was counted in 10 randomly selected fields using inverted light microscope at ×20 magnification, and all counts were normalized to the control.
Flow cytometry analysis
The levels of the CD44+, CD24 -, and CD133+cells in each group were evaluated using flow cytometry with a fluorescein isothiocyanate (FITC)-conjugated CD44 antibody (cat. no. 347943; BD Biosciences), phycoerythrin (PE)-conjugated CD24 antibody (cat. no. 555428; BD Biosciences), and a PE-conjugated CD133 antibody (cat. no. 12-1331-82; eBioscience; Thermo Fisher Scientific, Inc.). Mouse immunoglobulin (Ig)G2a-FITC (cat. no. 555573; BD Biosciences), mouse IgG2a-PE (cat. no. 554648; BD Biosciences), and mouse IgG1K-PE (cat. no. 12-4714-42; eBiosciences Thermo Fisher Scientific, Inc.) were used as the isotype controls (all of antibodies diluted at 1:100 ratio). Cells were incubated with antibodies at 4˚C for 45 minutes and analyzed using FACSCaliburTM (Becton Dickinson). The raw data were analyzed using Flowing Software version 2.5.0 (Perttu Terho).
Reverse transcription-quantitative (q-RT) PCR
Total RNA was extracted from mammospheres in control and treated groups using TRIzol® reagent (Sigma-Aldrich; Merck KGaA) and any DNA was digested using RNase free DNase I (Takara Bio, Inc.) according to the manufacturer’s protocol. cDNA was generated using RevertAid H Minus First Strand cDNA kit (Fermentas; Thermo Fisher Scientific, Inc.) according to the manufacturer’s instructions. The thermocycling conditions for qRT-PCR were: 95˚C for 15 seconds and 60˚C for 60 seconds for 40 cycles. The expression of the stemness-associated genes OCT4, c-MYC, KLF4, SOX2, CK8, CK18, CK19, NANOG, CDH1, CDH2, SNAIL1, SNAIL2, TWIST1, TWIST2, and ZEB1 was assessed using a Rotor Gene 6000 Real-Time PCR machine (Qiagen China Co., Ltd.) and analyzed using Rotor Gene 6000 version 1.7 (Qiagen China Co., Ltd.). SYBR Green PCR Master mix (Applied Biosystems) was used for qPCR according to the manufacturer’s protocol. The list of human-specific primers used, and their sequences are presented in Table 1. Melting curve analysis from 65-95˚C was performed, the results were normalized to GAPDH, and data were analyzed using the 2-∆∆Cq method.
29
Table 1.
Primer sequences used for reverse transcription-quantitative PCR
|
Primer
|
Sequence
|
Length, bp
|
NCBI accession number
|
| SOX2 |
F, 5’-GGGAAATGGAAGGGGTGCAAAAGAGG-3’
R, 5’-TTGCGTGAGTGTGGATGGGATTGGTG-3’
|
151 |
NM_003106.3 |
| Klf4 |
F, 5’-ATTACCAAGAGCTCATGCCA-3’
R, 5’-CCTTGAGATGGGAACTCTTTG-3’
|
150 |
NM_004235.4 |
| NANOG |
F, 5’-AAAGAATCTTCACCTATGCC-3’
R, 5’-GAAGGAAGAGGAGAGACAGT-3’
|
110 |
NM_024865.2 |
| OCT4 |
F, 5’-CTGGGTTGATCCTCGGACCT-3’
R, 5’-CACAGAACTCATACGGCGGG-3’
|
128 |
NM_002701.4 |
| c-MYC |
F, 5’-ACACATCAGCACAACTACG-3’
R, 5’-CGCCTCTTGACATTCTCC-3’
|
140 |
NM_002467 |
| TWIST1 |
F, 5’-CCAGGTACATCGACTTCCTC-3’
R, 5’-TCGTGAGCCACATAGCTG-3’
|
85 |
NM_000474.3 |
| TWIST2 |
F, 5’- GCGCAAGTGGAATTGGGATG-3’
R, 5’- CGGGTCTTCTGTCCGATGTC-3’
|
128 |
NM_001271893.4 |
| CDH1 |
F, 5’-CAGGAGTCATCAGTGTGGT-3’
R, 5’-GGAGGATTATCGTTGGTGTCAG-3’
|
150 |
NM_004360.3 |
| CDH2 |
F, 5’-GCCCAAGACAAAGAGACCC-3’
R, 5’-CTGCTGACTCCTTCACTGAC-3’
|
93 |
NM_001792.3 |
| SNAIL1 |
F, 5’-CCAGAGTTTACCTTCCAGCA-3’
R, 5’-GATGAGCATTGGCAGCGA-3’
|
101 |
NM_005985.3 |
| SNAIL2 |
F, 5’-AACTACAGCGAACTGGACAC-3’
R, 5’-GGATCTCTGGTTGTGGTATGAC-3’
|
90 |
NM_003068.3 |
| β-actin |
F, 5’-TCCCTGGAGAAGAGCTACG-3’
R, 5’-GTAGTTTCGTGGATGCCACA-3’
|
131 |
NM_ 001101.3 |
| ZEB1 |
F, 5’-GAGGATGACACAGGAAAGGA-3’
R, 5’-CAGCAGTGTCTTGTTGTTGT-3’
|
163 |
NM_0011281282 |
| NOTCH |
F, 5’-CAGACCCACACCCAGTA-3’
R, 5’-GGCAACGTCAACACCTT-3’
|
114 |
NM_017617 |
| CD133 |
F, 5’-GCATCCATCAAGTGAAACGT-3’
R, 5’-GGTTTGGCGTTGTACTCTGT-3’
|
199 |
NM_001145852.1 |
| CK8 |
F, 5’-CAGATCAAGACCCTCAACAAC-3’
R, 5’-CACTTGGTCTCCAGCATCTT-3’
|
89 |
NM_001256293.1 |
| CK18 |
F, 5’-GCGAGGACTTTAATCTTGGTG-3’
R, 5’-CTTTGGTGTCATTGGTC CAG-3’
|
120 |
NM_199187.1 |
| CK19 |
F, 5’-GCGACTACAGCCACTACTACA-3’
R, 5’-TGGTTCGGAAGTCATCTGC-3’
|
129 |
NM_002276.4 |
Generation of a BC mouse model
All studies in vivo were performed according to the guidelines for animal care established by the Royan Institute’s Animal Care Committee and approved by the Institutional Review Board and Ethics Committee of the Royan Institute. A total of 1×106 4T1 cells in 100 µL Matrigel were injected in the flank of inbred females Balb-C mice (n: 15 mice, weight: 18-21 g, Supplier: Royan Institute). The mice developed tumors 7 days after injection of cells, at which point, tumor-bearing mice were divided into four groups randomly: Control, solvent, 10X, and 20X. Each group contained three mice. The control group did not receive any treatment, the solvent group received ethanol, the 10X group received 1 mg/kg silibinin (10 fold dose of in vitro), and the 20X group received 2 mg/kg silibinin (20 fold dose of in vitro). The solvent and silibinin were injected directly into the tumor. Injections of solvent and silibinin were performed every 72 hours after the tumor diameter reached 4-6 mm, and tumor size was measured daily with calipers. All mice were sacrificed at day 20post-treatment and tumors were removed for pathological and immunohistochemical evaluation.
Hematoxylin and eosin (H& E) staining and immunohistochemistry
Tumor tissues were removed, fixed in 10% formalin at RT for 72 hours, and embedded into paraffin blocks. Tissues were sectioned (5 µm thickness), deparaffinized using xylol, and stained using H&E at RT for 45 minutes. The rate of mitosis, pleomorphism, inflammation, karyorrhexis, and desmoplasia per a field of view was evaluated by an independent pathologist at a magnification of ×400. For Immunohistochemistry evaluation, the slides were deparaffinized and rehydrated in a decreasing series of ethanol solutions (two incubations in 100% for 3 minutes each, 96% for 3 minutes, 70% for 3 minutes, and distilled water for 3 minutes). Slides were incubated in 3% hydrogen peroxide in methanol for 20 minutes at room temperature to inhibit endogenous peroxidase activity. Citrate buffer (pH = 6.0) and autoclaving for 10 minutes at 120˚C with pressure were used for antigen retrieval. After the slides had cooled, they were washed in Tris-buffered saline (TBS). The sections were incubated at 4˚C overnight with anti-human α-SMA antibody recognizing the cytoplasmic SMA which surrounded the vessels (1:200; Cat. No. ab7817; Abcam). The sections were washed the following day and incubated with the horseradish peroxidase-conjugated anti-mouse secondary antibody diluted in PBS - (1:500; Cat. No. ab205719; Abcam) for 60 minutes at RT. The sections were then stained with 3, 3’-diaminobenzidine (Dako) substrate as the chromogen for two minutes in the dark and at room temperature. Subsequently, the sections were counterstained with hematoxylin (Dako; Agilent Technologies, Inc.). All images were captured on an Olympus BX51 fluorescence microscope.
Western blot analysis
A total of 35 µg protein was extracted from treated and non-treated mammospheres using Q Proteom Mammalian Protein Prep kit (Cat. No. 37901; Thermo Fisher Scientific, Inc.), preheated at 100˚C for 3 minutes in a reducing sample buffer containing 50 mM Tris-Cl (pH 6.8), 2% SDS, 10% glycerol, 0.1% Bromophenol blue, and 100 mM β-Mercaptoethanol, were loaded on a 10% SDS gel and resolved using SDS-PAGE. Protein concentration was measured using a protein assay kit (cat. no. 23225; Thermo Fisher Scientific, Inc.). Proteins were subsequently transferred to a PVDF membrane (Whatman Plc; GE Healthcare Life Sciences). After transfer, membranes were blocked with 5% BSA in TBS at RT for 60 minutes, followed by an overnight incubation with either anti-phospho-STAT3 or anti-GAPDH (Sigma Aldrich; Merck KGaA; cat. no. ZRB1004) in 5% BSA in TBS at 4˚C (both 1:1,000). After washing with TBS containing 0.1% Tween 20, the membranes were incubated with horseradish peroxidase-conjugated secondary antibody (Abcam; cat. no. ab205719; 1:7,000) in 5% skimmed milk powder in TBS for 2 hours at RT. Protein bands were visualized using Pierce enhanced chemiluminescence western blotting substrate (Pierece; Thermo Fisher Scientific, Inc.; cat. no. 32106) and western blotting was performed three times for each experiment independently.
Search Tool for Interactions of Chemicals (STITCH)
STITCH (version 5; stitch.embl.de/) is a database of known and predicted interactions between chemicals and proteins based on computational predictions and established interactions in other organisms, and from interactions aggregated from other (primary) databases. The interactions include direct (physical) and indirect (functional) associations. The STITCH database was used and the experimentally validated genes and silibinin were searched. Using the STITCH database, interactions between silibinin and genes that were validated in the present study were evaluated.
Statistical analysis
Student’s t test, and one-way ANOVA or a two-way ANOVA were used for comparing the means of two independent groups, comparing more than 2 independent groups or analyzing and comparing the means of dependent groups, respectively. All experiments were performed in triplicate, and data were presented as the mean ± standard deviation. P ≤ 0.05 was considered to indicate a statistically significant difference. Statistical analysis was performed using GraphPad Prism version 5 (GraphPad Software, Inc.).
Results
Silibinin reduces the viability and growth rate of mammospheres
To determine the cytotoxic effect of silibinin on mammospheres, mammospheres were treated with different concentrations of silibinin for 72 hours. The results showed that silibinin significantly reduced cell viability of mammospheres in a dose-dependent manner (Fig. 1A-C). The IC50 dose of silibinin was determined to be 150 μM, 100 μM, and 50 μM for MCF-7, MDA-MB-231 derived-mammospheres, and MDA-MB-468 cell aggregation, respectively. Furthermore, mammospheres treated with the IC50 doses of silibinin, exhibited significantly reduced proliferation 120 hours post-treatment (P < 0.001; Fig. 1D-F). Mammospheres derived from MCF-7 cells were presented with the largest reduction in mammosphere growth when treated with silibinin.
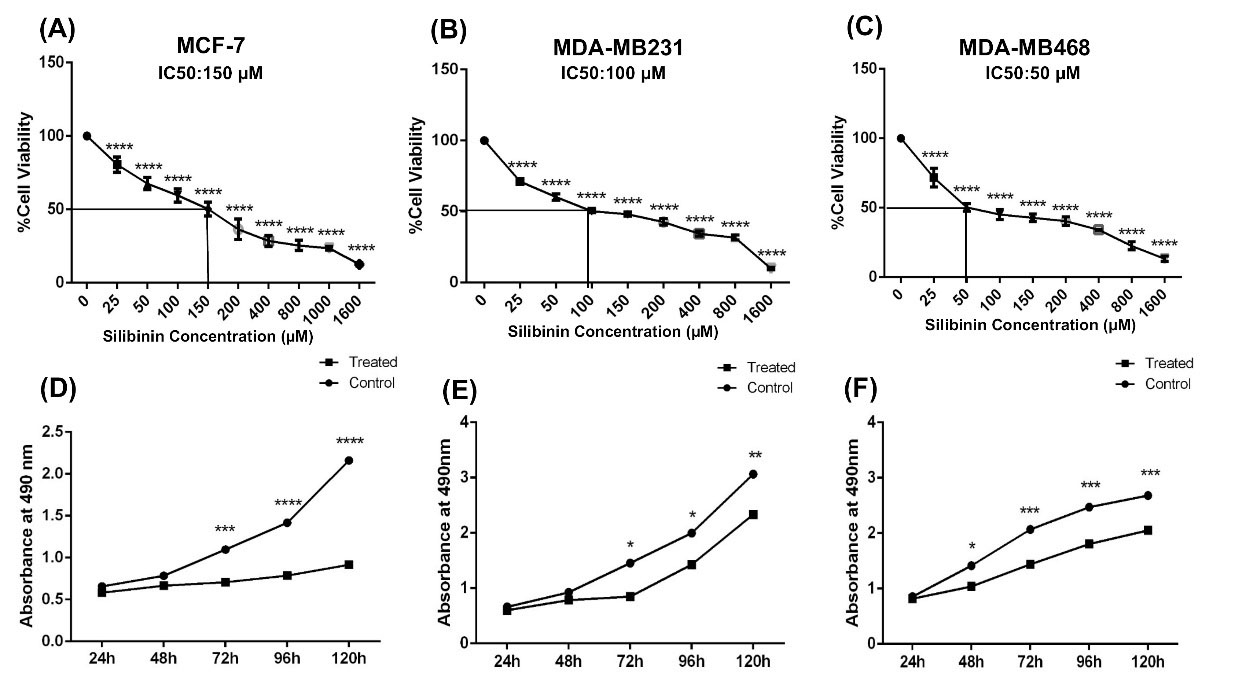
Fig. 1.
Silibinin reduces the viability and proliferation of mammospheres. Mammospheres derived from breast cancer lines were treated with 0-1600 µM of silibinin for 72 hours and the viability of cells were assessed. The IC50 doses of silibinin were determined to be (A) MCF-7, 150 µM; (B) MDA-MB-231, 100 µM; Mammospheres and (C) MDA-MB-468, 50 µM. (D-F) aggregated cells were treated with the respective IC50 doses and proliferation was assessed in cells treated for 24, 48, 72, 96 and 120 hours. Silibinin-treated cells exhibited significantly reduced proliferation compared with the respective controls at the same time point. Data are presented as the mean + standard deviation of three different biological repeats. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
.
Silibinin reduces the viability and proliferation of mammospheres. Mammospheres derived from breast cancer lines were treated with 0-1600 µM of silibinin for 72 hours and the viability of cells were assessed. The IC50 doses of silibinin were determined to be (A) MCF-7, 150 µM; (B) MDA-MB-231, 100 µM; Mammospheres and (C) MDA-MB-468, 50 µM. (D-F) aggregated cells were treated with the respective IC50 doses and proliferation was assessed in cells treated for 24, 48, 72, 96 and 120 hours. Silibinin-treated cells exhibited significantly reduced proliferation compared with the respective controls at the same time point. Data are presented as the mean + standard deviation of three different biological repeats. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Silibinin reduces stemness properties and promotes differentiation of cells in mammospheres
An important characteristic of CSCs is a capacity to self-renew. To determine whether MCF-7, MDA-MB-231, and MDA-MB-468 cells exhibited this capacity in vitro, colony and sphere formation assays were used. In the colony formation assay, CSCs, progenitors, and fully differentiated cells propagated and created colonies with special characteristics. In the sphere formation assay, CSCs were the only cells which grew in serum-free medium on a non-adhesive surface.
27,30,31
There was a significant reduction in the number of colonies and colony size when mammospheres were treated with silibinin (Fig. 2A-D). Additionally, MCF-7-mammospheres treated with silibinin did not form spheroids, and MDA-MB-231- mammosphere and MDA-MB-468-cell aggregation formed significantly fewer (Fig. 2E and F) and smaller spheroids (Fig. 2G). The effect of silibinin on spheroid formation was most prominent in MCF-7-mammospheres compared with the other cells.
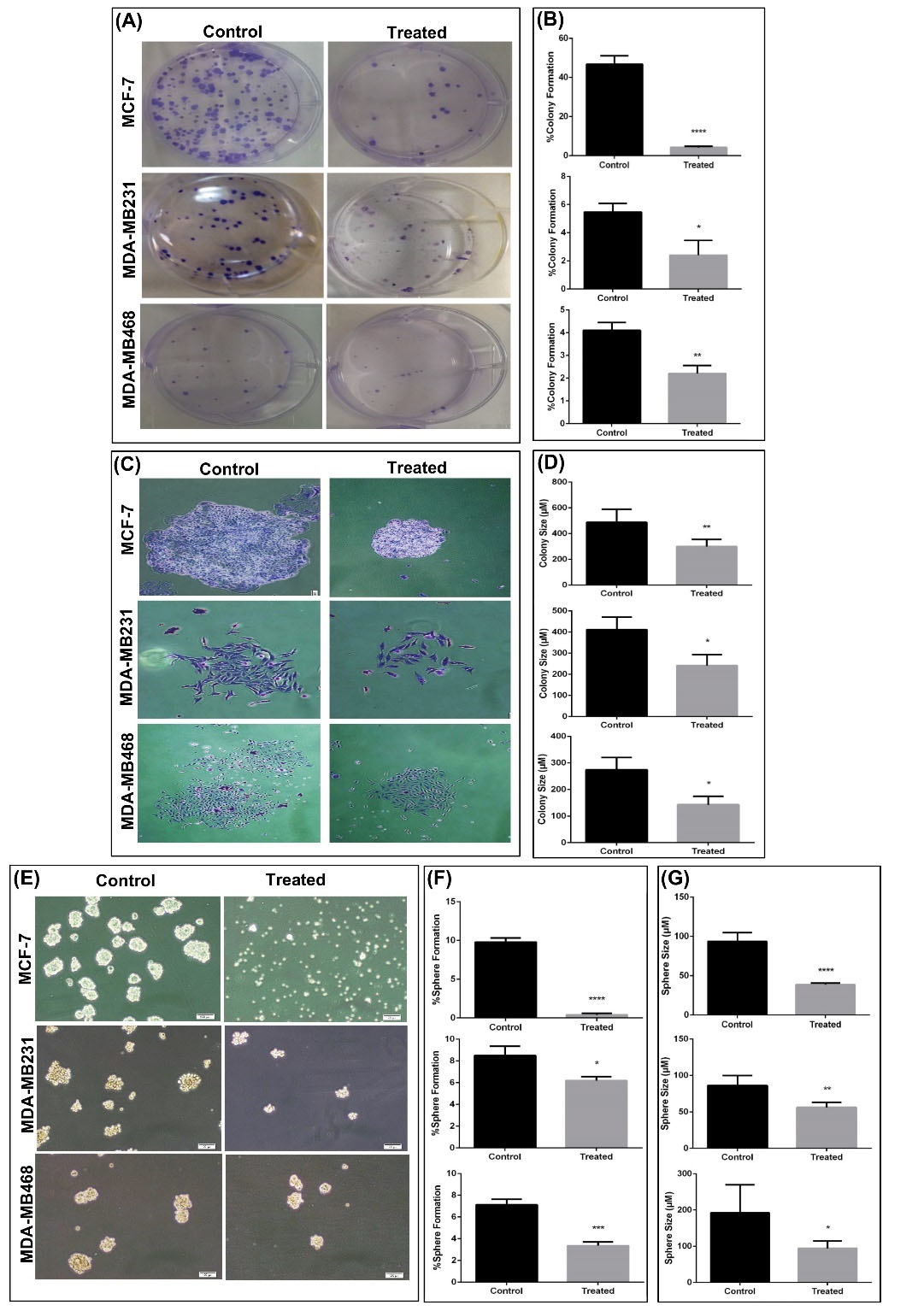

Fig. 2.
Colony and sphere formation ability is reduced in mammospheres treated with silibinin. (A) Representative images of colony formation in BC cell lines treated with control or the respective IC50 dose for 72 hours. (B) Percentage colony formation was calculated by counting the number of colonies in different cell lines and treatments. The number of colonies formed was significantly lower in the treated cells compared with the respective control. (C) Representative images of staining of a single colony with crystal violet. Magnification, x20. (D) The size of colonies were significantly smaller in cells treated with silibinin compared with the respective control. (E)Morphology of mammospheres and cell aggregation after 7 days of culture. (F) Sphere formation was significantly reduced in MCF-7 and MDA-MB-231 cell lines treated with silibinin compared with the respective untreated cells. The reduction in sphere formation was the largest in the MCF-7 cells. (G) Sphere size was significantly reduced in the cells treated with silibinin compared with the respective untreated cells. Data are presented as the mean + standard deviation of at least three biological repeats. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. BC, breast cancer.
.
Colony and sphere formation ability is reduced in mammospheres treated with silibinin. (A) Representative images of colony formation in BC cell lines treated with control or the respective IC50 dose for 72 hours. (B) Percentage colony formation was calculated by counting the number of colonies in different cell lines and treatments. The number of colonies formed was significantly lower in the treated cells compared with the respective control. (C) Representative images of staining of a single colony with crystal violet. Magnification, x20. (D) The size of colonies were significantly smaller in cells treated with silibinin compared with the respective control. (E)Morphology of mammospheres and cell aggregation after 7 days of culture. (F) Sphere formation was significantly reduced in MCF-7 and MDA-MB-231 cell lines treated with silibinin compared with the respective untreated cells. The reduction in sphere formation was the largest in the MCF-7 cells. (G) Sphere size was significantly reduced in the cells treated with silibinin compared with the respective untreated cells. Data are presented as the mean + standard deviation of at least three biological repeats. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. BC, breast cancer.
Based on the above results, the expression of stemness-associated genes, including OCT4, c-MYC, KLF4, and SOX2, and the differentiation-associated cytokeratin genes including CK8, CK18, and CK19 in mammospheres following treatment with silibinin was assessed. In treated mammospheres, expression of all the stemness-associated genes were all significantly downregulated except for c-MYC in MCF-7-mammospheres, and NANOG and KLF4 in MDA-MB-468 mammospheres (Fig. 3A-C). This reduction was accompanied with the overexpression of differentiation-associated genes in treated mammospheres derived from MDA-MB-231 and MDA-MB-468 (Fig. 3B and C). CK18 was upregulated in treated MCF-7- mammospheres (Fig. 3A). The STAT3 pathway is an active signaling pathway in tumors which activates and enhances cell proliferation,
28,32
and activity of this pathway has been suggested to be increased in BCSCs.
33,34
As shown in Fig. 3D, silibinin significantly reduced the levels of Stat3 phosphorylation at tyrosine 705 in MCF-7-mammospheres, although this was not altered in mammospheres based on the other two cell lines. However, it was not possible to measure the expression levels of total Stat3, thus it cannot be stated with certainty, whether the total levels of Stat3 were reduced or phosphorylation of Stat3 was reduced.
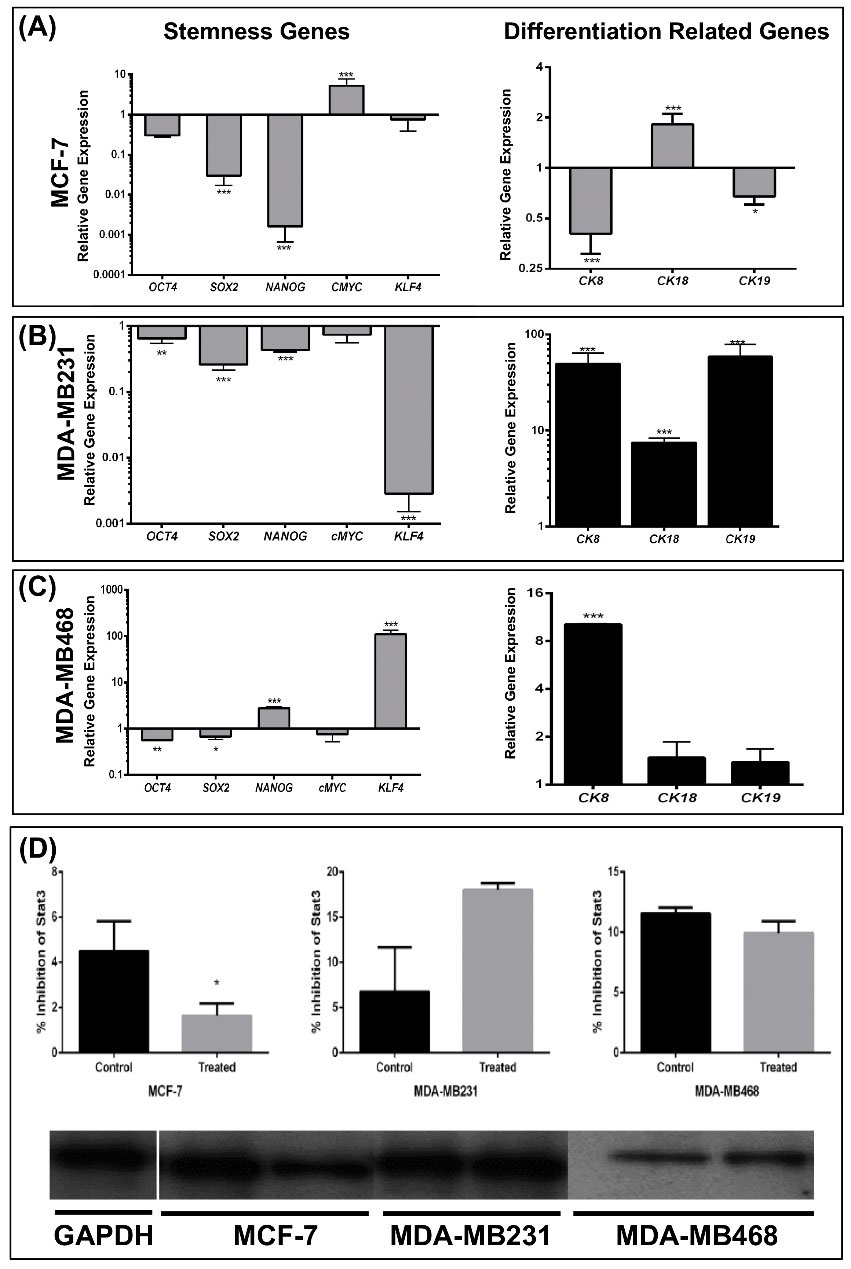

Fig. 3.
Silibinin modulates the expression of stemness and epithelial-associated cytokeratin genes in derived -mammospheres and aggregated cells. (A) mRNA expression levels of OCT4, SOX2, and KLF4 were reduced 2.4, 7.2, and 4.8 fold in treated MCF-7 mammospheres, respectively, and c-MYC was upregulated 7.4 fold in treated MCF-7 mammospheres. Expression of two differentiation-associated genes, CK8 and CK19, were significantly downregulated whereas CK18 was significantly upregulated. (B) Expression of stemness associated genes were all downregulated and all changes were determined to be significant except for c-Myc. All differentiation-associated genes were significantly upregulated in treated MDA-MB-231 cells. (C) In the MDA-MB-468 cells, expression of certain stemness-associated genes were upregulated whereas others were downregulated. NANOG and KLF4 exhibited the largest changes in expression and both were upregulated. c-MYC expression was not significantly different. Expression of CK8 was significantly increased whereas expression of CK18 and CK19 was not determined to be significantly different. GAPDH was used as the internal control. (D) Western blot analysis revealed that p-STAT3 protein expression was significantly reduced in the treated MCF-7 mammospheres and was not significantly changed in the other cell lines when treated with silibinin. However, the reduction in p-STAT3 may be the result of a reduction in total STAT3 expression which was not assessed. Data are presented as the mean ± standard deviation of three biological repeats. **P < 0.01, ***P < 0.001, ****P < 0.0001. p-, phospho; OD, optical density.
.
Silibinin modulates the expression of stemness and epithelial-associated cytokeratin genes in derived -mammospheres and aggregated cells. (A) mRNA expression levels of OCT4, SOX2, and KLF4 were reduced 2.4, 7.2, and 4.8 fold in treated MCF-7 mammospheres, respectively, and c-MYC was upregulated 7.4 fold in treated MCF-7 mammospheres. Expression of two differentiation-associated genes, CK8 and CK19, were significantly downregulated whereas CK18 was significantly upregulated. (B) Expression of stemness associated genes were all downregulated and all changes were determined to be significant except for c-Myc. All differentiation-associated genes were significantly upregulated in treated MDA-MB-231 cells. (C) In the MDA-MB-468 cells, expression of certain stemness-associated genes were upregulated whereas others were downregulated. NANOG and KLF4 exhibited the largest changes in expression and both were upregulated. c-MYC expression was not significantly different. Expression of CK8 was significantly increased whereas expression of CK18 and CK19 was not determined to be significantly different. GAPDH was used as the internal control. (D) Western blot analysis revealed that p-STAT3 protein expression was significantly reduced in the treated MCF-7 mammospheres and was not significantly changed in the other cell lines when treated with silibinin. However, the reduction in p-STAT3 may be the result of a reduction in total STAT3 expression which was not assessed. Data are presented as the mean ± standard deviation of three biological repeats. **P < 0.01, ***P < 0.001, ****P < 0.0001. p-, phospho; OD, optical density.
Subsequently, the expression levels of surface markers associated with BCSCs, including CD44+/CD24-
35
and CD133+CD44+
36,37
following silibinin treatment were assessed. CD44 is a membrane glycoprotein which acts as a receptor for hyaluronic acid, and participates in cell adhesion migration and metastasis.
38,39
However, CD24 as an adhesion molecule is upregulated during differentiation.
40
To determine the percentage of positive cells, the viable cell population were gated for using the forward scattering and side scattering results (Fig. 1 and S1). The percentage of CD44+/CD24 - cells were significantly lower in the MCF-7-mammospheres treated with silibinin compared with the respective control (Fig. 4A and 4B), and did not differ significantly in the other cell lines. The mean fluorescence intensity (MFI) of CD44+ cells were significantly increased in treated MDA-MB-231 and MDA-MB-486 cells. However, the MFI of CD44 in MDA-MB-468 was not significant when dual staining was done with CD133. MFI of CD44 did not change in the MCF-7 cells before and after Silibinin treatment (Fig. 4C and S2). CD133 expression was not altered by treatment with silibinin in the MDA-MB-231 and MDA-MB-468 cells, whereas expression was significantly reduced in the MCF-7 mammospheres when treated (Fig. 4D-F).
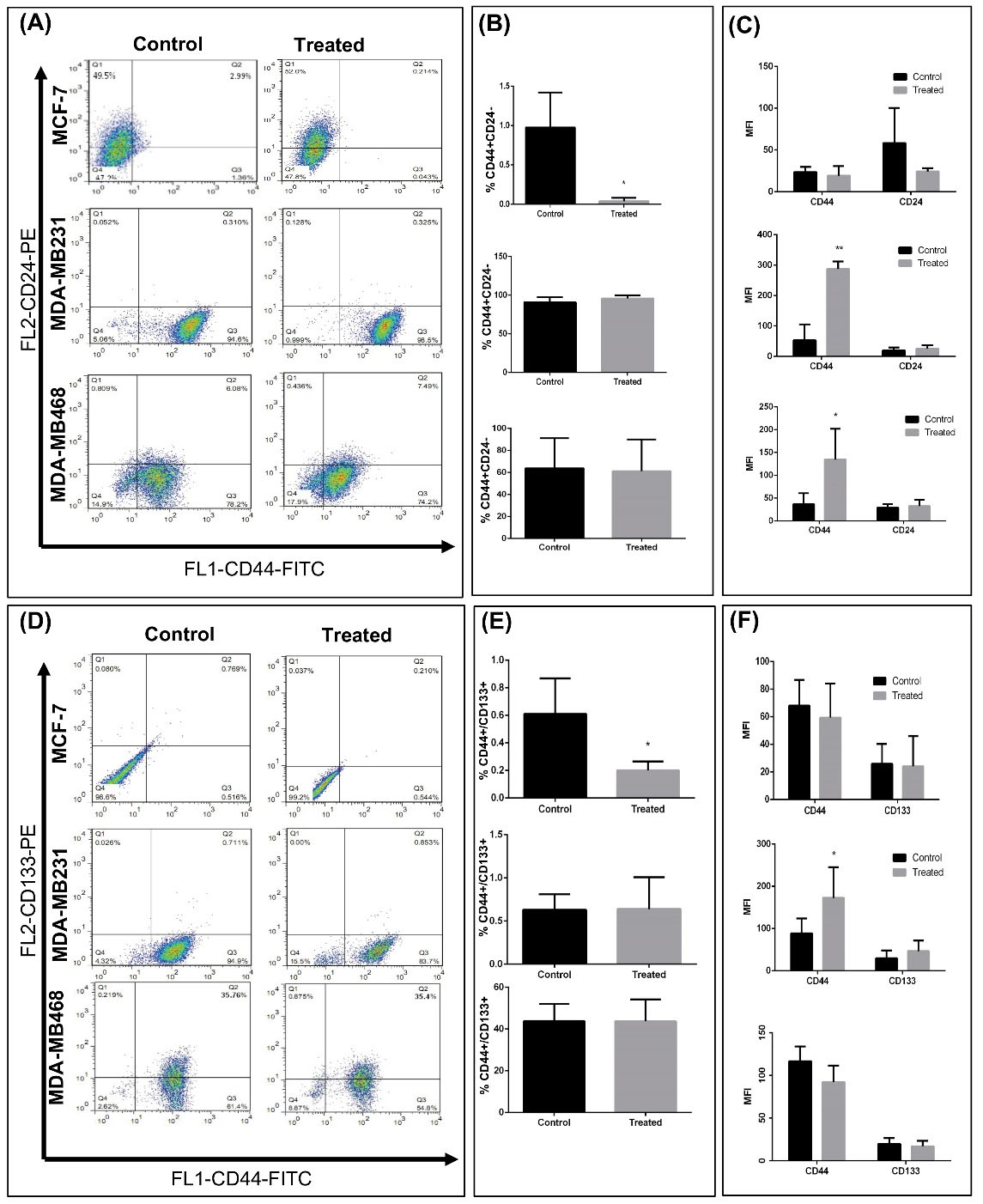

Fig. 4.
Evaluation of breast cancer stem cell markers in mammosphere cells treated with silibinin. (A) Representative flow cytometry dot plots of CD24/CD44 stem cell markers in cells obtained from mammospheres. (B) Quantitative analysis of CD44/CD24 surface markers expression in treated and untreated mammospheres. A significant decrease was observed in CD44+/CD24 - cells in treated MCF-7 cells. (C) MFI of CD44 and CD24 in each group. CD44 intensity increased in treated derrived –mammospheres from MD-MB231 and MDA-MB-468 aggregated cells. (D) Representative flow cytometry dot plots of CD133/CD44 markers in cells obtained from mammospheres. (B) Quantitative analysis of CD133/CD44 surface markers expression in treated and untreated mammospheres. A significant decrease was observed in CD44+/CD133+ cells in treated MCF-7 cells. (C) MFI of CD44 and CD133 in each group. CD44 intensity increased in treated MD-MB231 mammospheres. Data are presented as the mean ± standard deviation of three biological repeats. *P < 0.05, **P < 0.01 vs. control. MFI, mean fluorescence intensity; FITC, fluorescein isothiocyanate; PE, phycoerythrin., FL2, fluorescence parameter 2.
.
Evaluation of breast cancer stem cell markers in mammosphere cells treated with silibinin. (A) Representative flow cytometry dot plots of CD24/CD44 stem cell markers in cells obtained from mammospheres. (B) Quantitative analysis of CD44/CD24 surface markers expression in treated and untreated mammospheres. A significant decrease was observed in CD44+/CD24 - cells in treated MCF-7 cells. (C) MFI of CD44 and CD24 in each group. CD44 intensity increased in treated derrived –mammospheres from MD-MB231 and MDA-MB-468 aggregated cells. (D) Representative flow cytometry dot plots of CD133/CD44 markers in cells obtained from mammospheres. (B) Quantitative analysis of CD133/CD44 surface markers expression in treated and untreated mammospheres. A significant decrease was observed in CD44+/CD133+ cells in treated MCF-7 cells. (C) MFI of CD44 and CD133 in each group. CD44 intensity increased in treated MD-MB231 mammospheres. Data are presented as the mean ± standard deviation of three biological repeats. *P < 0.05, **P < 0.01 vs. control. MFI, mean fluorescence intensity; FITC, fluorescein isothiocyanate; PE, phycoerythrin., FL2, fluorescence parameter 2.
Silibinin reduces the invasive and migratory potential of mammospheres
It has been hypothesized that CSCs potentiate invasion of cancer cells
41
Therefore, the effect of silibinin on invasion and migration of mammospheres was determined. As shown in Fig. 5, both the migratory and invasive capacity of mammospheres was reduced significantly following treatment with silibinin at the respective IC50 doses. The number of cells which had invaded decreased 4.6 fold (P < 0.0008) and the number cells which had migrated decreased 1.3-fold in the MCF-7 mammospheres (Fig. 5A), 4.2-fold decrease (invasion) and 2-fold decrease (migration) in the MDA-MB-231 mammospheres (Fig. 5B), and 2-fold decrease (invasion) and 3-fold decrease (migration) in MDA-MB-468 cells when treated with silibinin (Fig. 5C). Reduction of invasion and migration in MDA-MB-231 and MDA-MB-468 cells were associated with downregulation of EMT-associated genes (Fig. 5B and 5C). In MCF-7 mammospheres, several EMT-associated genes, including SNAIL2 and TWIST1/2 were upregulated when treated with silibinin, although the migratory and invasive capacity were decreased significantly (Fig. 5).
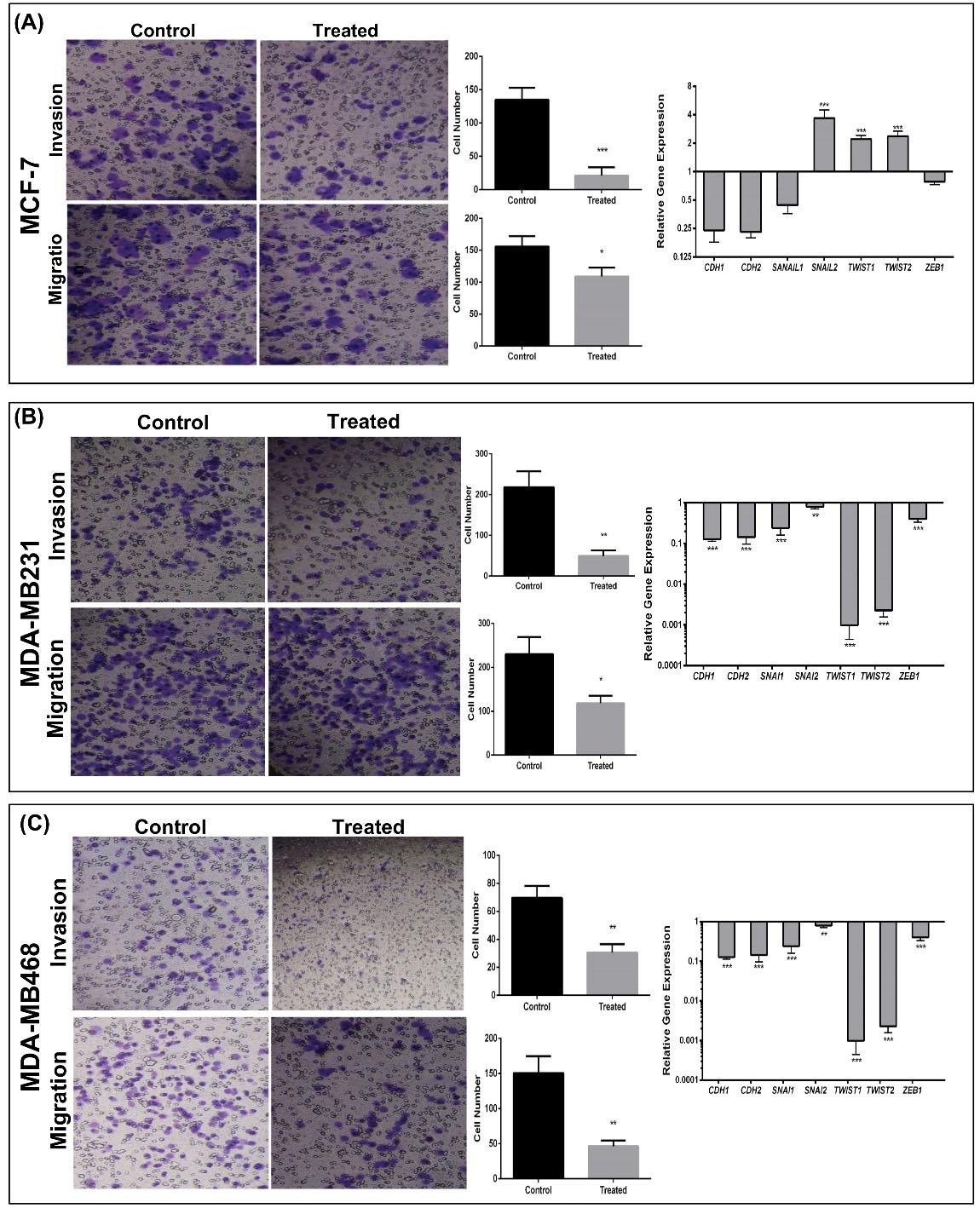
Fig. 5.
Invasive and migratory capacity of silibinin-treated cells from mammospheres. (A-C) The migratory and invasive capacities of mammospheres were reduced in all silibinin-treated cells. These reductions were accompanied by downregulation of important transcription factors known to regulate metastasis in MDA-MB-231 and MDA-MB-468 cells, whereas treated MCF-7 cells exhibited upregulation of some transcription factors. (Left) Migrated and invaded cells visualized with crystal violet. (Center) Quantitative analysis of the number of migrated (upper) and invaded (lower) cells. (Right) Expression of epithelial-mesenchymal transition associated genes in treated compared with untreated mammospheres. Expression of genes was normalized to GAPDH. Data are presented as the mean ± standard deviation of three biological repeats. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control.
.
Invasive and migratory capacity of silibinin-treated cells from mammospheres. (A-C) The migratory and invasive capacities of mammospheres were reduced in all silibinin-treated cells. These reductions were accompanied by downregulation of important transcription factors known to regulate metastasis in MDA-MB-231 and MDA-MB-468 cells, whereas treated MCF-7 cells exhibited upregulation of some transcription factors. (Left) Migrated and invaded cells visualized with crystal violet. (Center) Quantitative analysis of the number of migrated (upper) and invaded (lower) cells. (Right) Expression of epithelial-mesenchymal transition associated genes in treated compared with untreated mammospheres. Expression of genes was normalized to GAPDH. Data are presented as the mean ± standard deviation of three biological repeats. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control.
Silibinin reduces tumor growth in vivo
To determine the effect of silibinin in vivo, a BC mouse model was used. Treatment with silibinin significantly reduced tumor volume after 3 days (Fig. 6A) and tumor weight (Fig. 6B). H&E staining showed a notable reduction in proliferation in mice treated with both 10- and 20-fold doses of silibinin (Fig. 6C). The infiltration of polymorphonuclear cells, inflammation, and necrosis was also increased in the treated mice (Fig. 6C; Table 2). Furthermore, a notable reduction of α-SMA staining of endothelial tubes was observed in the mice treated with silibinin (Fig. 6D), suggesting that angiogenesis was reduced.
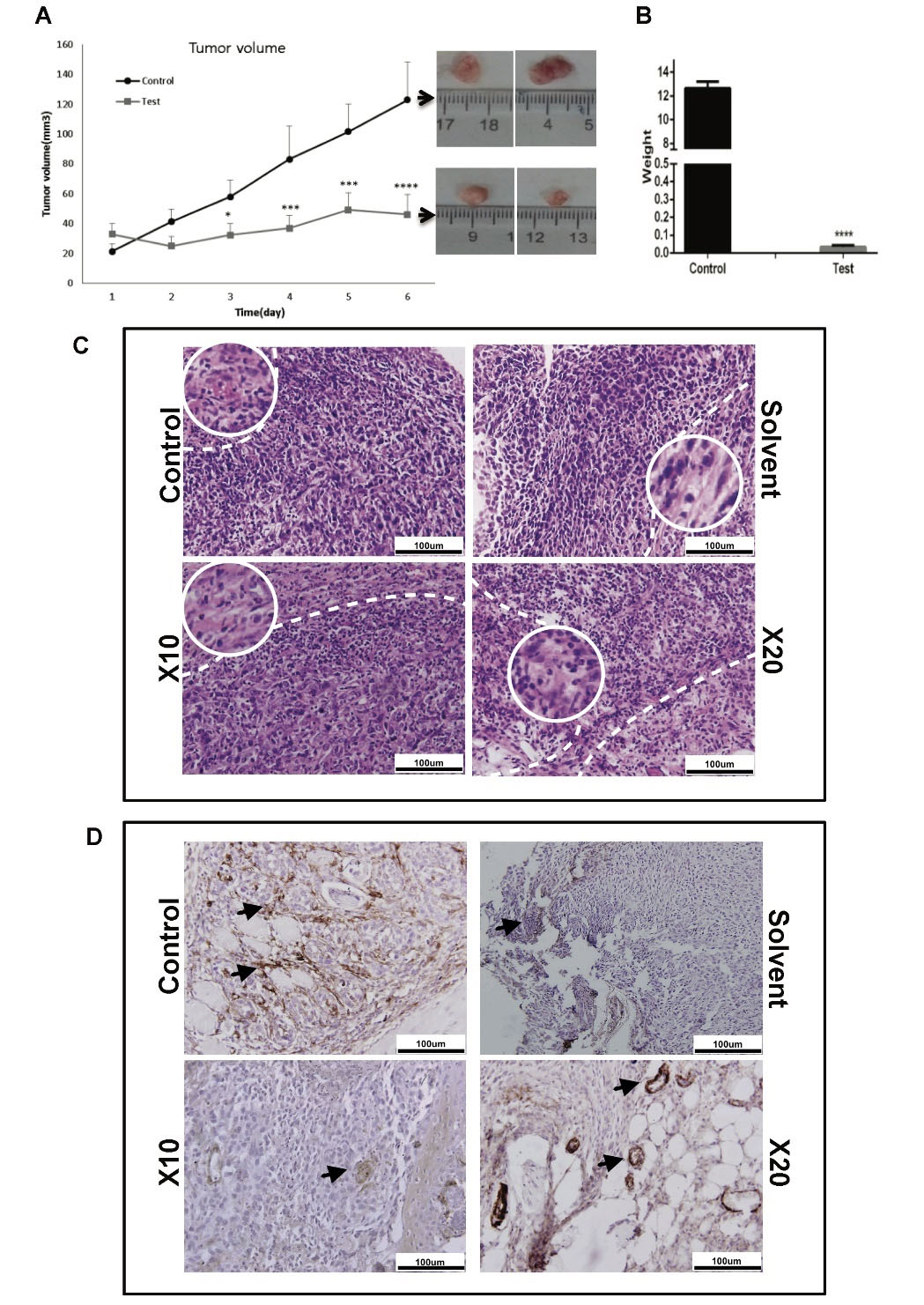
Fig. 6.
The effect of silibinin on tumor growth in a breast cancer mouse model. Silibinin (2 mg/kg) was injected intra-tumor every 48 hours in mice exhibiting tumors with a 4-6 mm diameter. (A) Tumor volume and (B) weight were significantly reduced in treated mice compared with the untreated group. (C) Hematoxylin and eosin staining of tumors showed an apparent increase in the number of necrotic cells in the silibinin-treated group. Scale bar, 100 µM (10X magnification). The white dash line showed necrotic area in each group and white circle indicated necrotic cell morphology in 20X-enlarged region. (D) Immunostaining for α-SMA. Expression of α-SMA (black arrows) was decreased in the tumors of treated mice. Scale bar, 100 µM.
.
The effect of silibinin on tumor growth in a breast cancer mouse model. Silibinin (2 mg/kg) was injected intra-tumor every 48 hours in mice exhibiting tumors with a 4-6 mm diameter. (A) Tumor volume and (B) weight were significantly reduced in treated mice compared with the untreated group. (C) Hematoxylin and eosin staining of tumors showed an apparent increase in the number of necrotic cells in the silibinin-treated group. Scale bar, 100 µM (10X magnification). The white dash line showed necrotic area in each group and white circle indicated necrotic cell morphology in 20X-enlarged region. (D) Immunostaining for α-SMA. Expression of α-SMA (black arrows) was decreased in the tumors of treated mice. Scale bar, 100 µM.
Table 2.
Histopathological analysis of tumor tissues in different groups
|
Group name
|
Mitosis per HPF
|
Pleomorphism
|
Inflammation
|
Karyorrhexis
|
Desmoplasia
|
Other
|
| Control |
6-7 |
2 |
Mild lymphocytic |
Not determined |
No |
Lymph node involving fat |
| Solvent |
6-7 |
2 |
Mild lymphocytic |
Not determined |
No |
- |
| X 10 |
2-3a
|
2 |
Moderate to severe lymph/PMN |
+ |
No |
Necrosis ++ |
| X 20 |
2-3a
|
2 |
Severe lymph/PMN |
+ |
No |
Necrosis + |
a
P < 0.05. HPF, high power fields; PMN, polymorphonuclear neutrophils; Control, tumor bearing mice without any treatment; solvent, tumor bearing mice that received; X10, tumor bearing mice that received 1 mg/kg silibinin every 48 hours. X20, tumor bearing mice that received 2 mg/kg silibinin every 48 hours; +/++, quality observation.
Silibinin interacts directly with genes which contribute to metastasis and indirectly to self-renewal transcription factors
As shown in Fig. 7A, silibinin directly interacted with ZEB1, SNAIL1, CDH1, and CTNBB1, and indirectly interacted with a number of major nodes, including the majority of the self-renewal and EMT associated genes. The catenin family (CTNN) (cadherin-associated proteins) formed large nodes in the identified network (Fig. 7B).
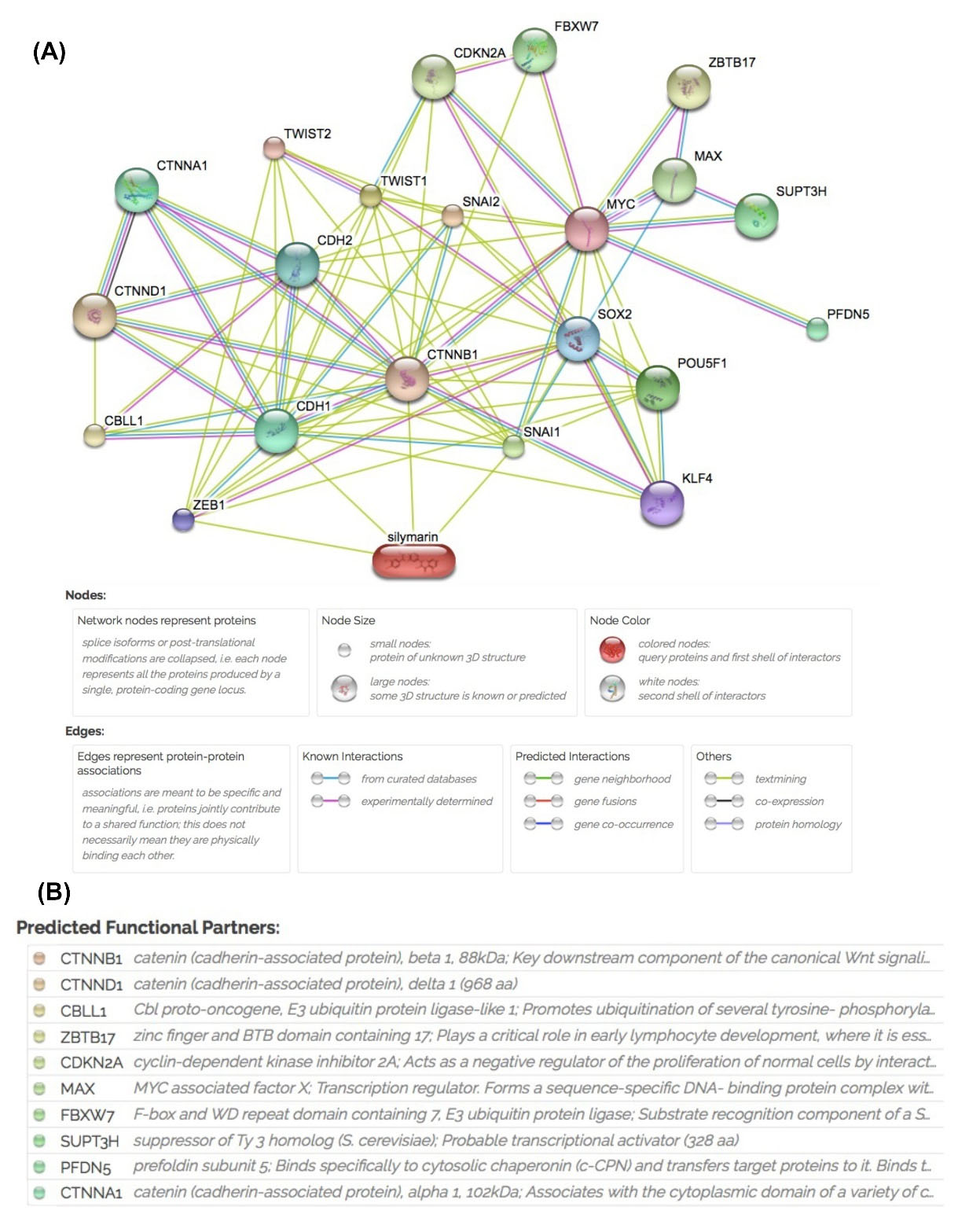
Fig. 7.
Search Tool for Interactions of Chemicals analysis of silibinin interactions with proteins important in self-renewal and metastasis. (A) The network shows different nodes with various interactions between silibinin and associated genes including predicted interactions. (B) Predicted functional partner nodes and their function.
.
Search Tool for Interactions of Chemicals analysis of silibinin interactions with proteins important in self-renewal and metastasis. (A) The network shows different nodes with various interactions between silibinin and associated genes including predicted interactions. (B) Predicted functional partner nodes and their function.
Discussion
The aim of the present study was to determine whether silibinin, which reduces tumor viability, could target BC stem cells. In our previous study, it was demonstrated that silibinin exhibited anti-cancer effects by inhibiting the effects of anti-apoptotic genes, including Bcl-2 and Survivin,
27
reducing tumor volume in 3D cultures, reducing sphere formation, and reducing colony formation. However, the effects of silibinin on BC stem cells were unknown. Mammospheres were used in the present study as mammospheres are rich in CSCs,
42-44
and the presence of a rare population of CSCs was confirmed based on an immunophenotyping model using CD44, CD24, and CD133 antibodies. Based on our previous study using other cell lines and on other published studies,
45,46
cell surface markers cannot be used as the only criteria for proper characterization of CSCs. Spheroid culture conditions may be a more effective approach to enrich CSCs, rather than the use of cell surface markers.
30,31
Additionally, mammospheres were an appropriate model for the present study as they simulate the geometric, mechanical, and biochemical aspects of a tumor more accurately, and they better replicate cell-cell, and cell-substrate interactions, which in turn regulate proliferation and differentiation.
30,31
Mammospheres were derived from two different types of BC cell lines; MCF-7 (which were used as an ER+, PR+ cell line), and MDA-MB-231 (which were both used as triple negative BC cell lines). Mammospheres were treated with silibinin and the effects of treatment on expression of stemness, differentiation-associated, and EMT associated genes, and migration and invasion were determined. The IC50 of silibinin for MCF-7-mammospheres was 150 μM, for MDA-MB-231 was 100 μM and for MDA-MB-468-mammospheres was 50 μM. At the respective IC50 doses, colony formation, sphere formation, and proliferation were all reduced, and the reduction was the largest in the MCF-7 cells. These results are consistent with our previous study,
47
and other reports in ovarian,
18
lung,
45
and colon cancer.
46
Treatment with silibinin reduced self-renewal potential, and this was associated with a reduction in mammosphere formation, proliferation, and colony formation. The proportion of CD44+/CD24 - and CD44+/CD133+ cells, which are considered CSCs in BC,
48-50
were reduced in the treated MCF-7-mammospheres. However, the expression of the majority of stemness-associated genes was downregulated in all cell lines assessed, and the expression of differentiation-associated genes (CK8, CK18, and CK19) were upregulated in MDA-MB-231 and MDA-MB-468 cells when treated with silibinin. Similar results have been reported in other types of cancer, including head and neck,
51
colon,
25
nasopharyngeal carcinoma,
52
and colorectal cancer.
53
The expression of c-MYC and NANOG/KLF4 was upregulated in treated derived -mammospheres from MCF-7 cells and MDA-MB-468 aggregation cells, respectively. KLF-4 promotes or inhibits progression of cancer depending on the type of cancer and possibly other factors in the tumor microenvironment.
53-55
BC cell lines treated with silibinin exhibited upregulated expression of CK8, CK18, and CK19, suggesting that certain sub-populations of cancer cells may have undergone EMT.
54-56
Therefore, upregulation of these cytokeratins in the present study may inhibit micro-metastasis. The STAT3 signaling pathway serves as an important pathway in increasing proliferation and promoting the self-renewal capacity through OCT4 or WNT signaling pathways in cancer.
57,58
Based on the results of the study, treatment with silibinin reduced the levels of phosphorylated STAT3 in MCF-7-mammospheres, although it is not certain whether the total levels of Stat3 were reduced. The decreased levels of phosphorylated STAT3 may underlie the reduction in cell proliferation observed in treated cells. It has also been reported that silibinin may affect the expression of genes involved in drug resistance, including AKT and ABCG2 through regulation of stemness factors
19
All alterations in cancer cell function were associated with a reduction in the tumorigenicity of BC cells in nude mice. Silibinin not only reduced proliferation and self-renewal in vivo, but also appeared to inhibit angiogenesis which has significant effects on tumor growth. α-SMA expression analysis was performed to determine differences in angiogenesis.
One limitation of the present study was an inaccessibility to Ki67 and VEGFantibodiesto confirm the results. The pattern of responses to silibinin in the two triple-negative BC cell lines were similar, and were both more sensitive to silibinin than the MCF-7 cells. The mechanism by which silibinin exhibits its effects are unknown, but it has been reported that silibinin inhibits progression of colorectal cancer stem-like cells through inhibition of the PP2A/AKT/mTOR signaling pathway,
46
as well as inhibiting the growth of colonospheres through the IL4/6 signaling pathway.
25
Furthermore, in human GBM, silibinin induced apoptosis and autophagy through simultaneous inhibition of the mTOR and YAP signaling pathways
59
; whereas in gastric cancer, silibinin reduced metastasis through the MAPK signaling pathway.
60
In the present study, a reduction in the migratory and invasive capacity of mammospheres formed from all BC cell lines was demonstrated when treated with silibinin. The reduction in invasiveness may have been associated with downregulation of certain EMT-transcription factors in MDA-MB-231 and MDA-MB-468 cells. However, SNAIL1 and TWIST1/2 expressions were upregulated in treated MCF-7 cells, but the migratory and invasive capacity were still significantly decreased. Therefore, silibinin may control both stemness and metastatic potential, and the balance of transcription factors by an unknown regulatory network which may determine the fate of cancer cells. Use of the STITCH database identified that silibinin may have exerted its effect directly by regulation of metastasis-associated genes, particularly ZEB1, SNAIL1, CDH1, and CTNBB1. Therefore, it is hypothesized that the direct effects of silibinin on metastasis-associated genes, particularly ZEB1, SNAIL1, CDH1, and CTNBB1, and the indirect effects on stemness-associated genes may be the mechanism by which silibinin affected cell behaviors. Although no signs of metastasis were observed in the treated and control group in vivo (data not shown), an additional control in the in vivo experiments should be used in future experiments to determine the effect of silibinin on metastasis in the mouse model.
Conclusion
In conclusion, it was demonstrated that silibinin significantly decreased mammosphere and aggregated cells viability, colony and sphere formation, and migration and invasion. Furthermore, silibinin altered differentiation in BC cells, highlighting its potential as a therapeutic option for treatment of BC.
Research Highlights
What is the current knowledge?
√ Silibinin has anti-tumor effects on various cancers.
√ Sphere-forming culture due to the availability of 3D culture is considered as a suitable alternative to animal models.
√ Sphere-forming culture is a proper method to CSCs enrichment.
What is new here?
√ Silibinin can affect various CSCs.
√ Anti-tumor effect of silibinin in breast cancer is related to direct targeting BCSCs.
√ Silibinin can reduce the tumorgenicity and metastasis and promote differentiation in mammospheres.
Acknowledgments
The authors would like to thank Mr. Payam Taheri, Mr. Abolfazle Kheimeh and Mr. Shahab Mirshahvaladi (Royan Institute for Stem Cell Biology and Technology, ACECR, Tehran, Iran) for their technical assistance and helpful advice.
Funding sources
The present study was funded by grants from Iran National Science Foundation (Grant no. 90003728) and Iranian Council of Stem Cell Research and Technology (Grant no. REP191).
Ethical statement
All procedures in the present study were performed in accordance with the relevant guidelines and regulations of the Royan Institute for Stem Cell Biology and Technology and approved by the Institutional Review Board and Ethics Committee of the Royan Institute, Tehran, Iran (approval no. IR.ACECR.ROYAN.REC.1395.35).
Competing interests
The authors declare that they have no competing interests.
Authors’ contribution
JF contributed to the conceptualization of the study, and data collection, and was a major contributor to writing the manuscript. AS, MR, PS, MA, and EJ contributed to data collection and data analysis. FS revised the manuscript and supervised the study. NS contributed to the pathological analysis. MS contributed to writing and editing the manuscript. ME contributed to the conceptualization of the study, revised the manuscript, and supervised the study. All authors read and approved the final manuscript.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Materials
Supplementary file 1 contains Figs. S1-S2.
(pdf)
References
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015; 136:E359-E86. doi: 10.1002/ijc.29210 [Crossref] [ Google Scholar]
- Keller PJ, Arendt LM, Skibinski A, Logvinenko T, Klebba I, Dong S. Defining the cellular precursors to human breast cancer. Proc Natl Acad Sci U S A 2012; 109:2772-7. doi: 10.1073/pnas.1017626108 [Crossref] [ Google Scholar]
- Vinogradov S, Wei XJN. Cancer stem cells and drug resistance: the potential of nanomedicine. Nanomedicine (Lond) 2012; 7:597-615. doi: 10.2217/nnm.12.22 [Crossref] [ Google Scholar]
- Chen K, Huang Y-h, Chen J-lJAPS. Understanding and targeting cancer stem cells: therapeutic implications and challenges. Acta Pharmacol Sin 2013; 34:732-40. doi: 10.1038/aps.2013.27 [Crossref] [ Google Scholar]
- Han L, Shi S, Gong T, Zhang Z, Sun XJAPSB. Cancer stem cells: therapeutic implications and perspectives in cancer therapy. Acta Pharmaceutica Sinica B 2013; 3:65-75. doi: 10.1016/j.apsb.2013.02.006 [Crossref] [ Google Scholar]
- Reynolds DS, Tevis KM, Blessing WA, Colson YL, Zaman MH, Grinstaff MWJSr. Breast cancer spheroids reveal a differential cancer stem cell response to chemotherapeutic treatment. Sci Rep 2017; 7:1-12. doi: 10.1038/s41598-017-10863-4 [Crossref] [ Google Scholar]
- Aggarwal BB, Danda D, Gupta S, Gehlot PJBp. Models for prevention and treatment of cancer: problems vs promises. Biochem Pharmacol 2009; 78:1083-94. doi: 10.1016/j.bcp.2009.05.027 [Crossref] [ Google Scholar]
- Friedrich J, Eder W, Castaneda J, Doss M, Huber E, Ebner R. A reliable tool to determine cell viability in complex 3-d culture: the acid phosphatase assay. J Biomol Screen 2007; 12:925-37. doi: 10.1177/1087057107306839 [Crossref] [ Google Scholar]
- Abbott A. Biology's new dimension. Nature 2003; 424:870-2. doi: 10.1038/424870a [Crossref] [ Google Scholar]
- Zhu X-X, Ding Y-H, Wu Y, Qian L-Y, Zou H, He QJErocp. Silibinin: a potential old drug for cancer therapy. Expert Rev Clin Pharmacol 2016; 9:1323-30. doi: 10.1080/17512433.2016.1208563 [Crossref] [ Google Scholar]
- Kaur M, Velmurugan B, Tyagi A, Deep G, Katiyar S, Agarwal C. Silibinin suppresses growth and induces apoptotic death of human colorectal carcinoma LoVo cells in culture and tumor xenograft. Mol Cancer Ther 2009; 8:2366-74. doi: 10.1158/1535-7163.MCT-09-0304 [Crossref] [ Google Scholar]
- Zhang X, Liu J, Zhang P, Dai L, Wu Z, Wang L. Silibinin induces G1 arrest, apoptosis and JNK/SAPK upregulation in SW1990 human pancreatic cancer cells. Oncol Lett 2018; 15:9868-76. doi: 10.3892/ol.2018.8541 [Crossref] [ Google Scholar]
- Deep G, Singh R, Agarwal C, Kroll D, Agarwal RJO. Silymarin and silibinin cause G1 and G2–M cell cycle arrest via distinct circuitries in human prostate cancer PC3 cells: a comparison of flavanone silibinin with flavanolignan mixture silymarin. Oncogene 2006; 25:1053-69. doi: 10.1038/sj.onc.1209146 [Crossref] [ Google Scholar]
- Surai PFJA. Silymarin as a natural antioxidant: an overview of the current evidence and perspectives. Antioxidants (Basel) 2015; 4:204-47. doi: 10.3390/antiox4010204 [Crossref] [ Google Scholar]
- Yang S-H, Lin J-K, Chen W-S, Chiu J-HJJoSR. Anti-angiogenic effect of silymarin on colon cancer LoVo cell line. J Surg Res 2003; 113:133-8. doi: 10.1016/s0022-4804(03)00229-4 [Crossref] [ Google Scholar]
- Loguercio C, Festi DJWjogW. Silybin and the liver: from basic research to clinical practice. World J Gastroenterol 2011; 17:2288. doi: 10.3748/wjg.v17.i18.2288 [Crossref] [ Google Scholar]
- Ramasamy K, Agarwal RJCl. Multitargeted therapy of cancer by silymarin. Cancer Lett 2008; 269:352-62. doi: 10.1016/j.canlet.2008.03.053 [Crossref] [ Google Scholar]
- Momeny M, Ghasemi R, Valenti G, Miranda M, Zekri A, Zarrinrad G. Effects of silibinin on growth and invasive properties of human ovarian carcinoma cells through suppression of heregulin/HER3 pathway. Tumour Biol 2016; 37:3913-23. doi: 10.1007/s13277-015-4220-6 [Crossref] [ Google Scholar]
- Deep G, Agarwal RJC, Reviews M. Antimetastatic efficacy of silibinin: molecular mechanisms and therapeutic potential against cancer. Cancer Metastasis Rev 2010; 29:447-63. doi: 10.1007/s10555-010-9237-0 [Crossref] [ Google Scholar]
- Liakopoulou C, Kazazis C, Vallianou NGJA-CAiMC. Silimarin and Cancer. Anticancer Agents Med Chem 2018; 18:1970-4. doi: 10.2174/1871520618666180905154949 [Crossref] [ Google Scholar]
- Chatran M, Pilehvar-Soltanahmadi Y, Dadashpour M, Faramarzi L, Rasouli S, Jafari-Gharabaghlou D. Synergistic anti-proliferative effects of metformin and silibinin combination on T47D breast cancer cells via hTERT and cyclin D1 inhibition. Drug Res (Stuttg) 2018; 68:710-6. doi: 10.1055/a-0631-8046 [Crossref] [ Google Scholar]
- Zhang Y, Ge Y, Ping X, Yu M, Lou D, Shi WJMmr. Synergistic apoptotic effects of silibinin in enhancing paclitaxel toxicity in human gastric cancer cell lines. Mol Med Rep 2018; 18:1835-41. doi: 10.3892/mmr.2018.9129 [Crossref] [ Google Scholar]
- Wu K, Ning Z, Zeng J, Fan J, Zhou J, Zhang T. Silibinin inhibits β-catenin/ZEB1 signaling and suppresses bladder cancer metastasis via dual-blocking epithelial–mesenchymal transition and stemness. Cell Signal 2013; 25:2625-33. doi: 10.1016/j.cellsig.2013.08.028 [Crossref] [ Google Scholar]
- Alimoghaddam K, Ghavamzadeh AJAoIm. Silibinin induces apoptosis and inhibits proliferation of estrogen receptor (ER)-negative breast carcinoma cells through suppression of nuclear factor kappa B activation. Arch Iran Med 2014; 17:366. [ Google Scholar]
- Kumar S, Raina K, Agarwal C, Agarwal RJO. Silibinin strongly inhibits the growth kinetics of colon cancer stem cell-enriched spheroids by modulating interleukin 4/6-mediated survival signals. Oncotarget 2014; 5:4972. doi: 10.18632/oncotarget.2068 [Crossref] [ Google Scholar]
- Mao J, Yang H, Cui T, Pan P, Kabir N, Chen D. Combined treatment with sorafenib and silibinin synergistically targets both HCC cells and cancer stem cells by enhanced inhibition of the phosphorylation of STAT3/ERK/AKT. Eur J Pharmacol 2018; 832:39-49. doi: 10.1016/j.ejphar.2018.05.027 [Crossref] [ Google Scholar]
- Abdollahi P, Ebrahimi M, Motamed N, Samani FSJA-cd. Silibinin affects tumor cell growth because of reduction of stemness properties and induction of apoptosis in 2D and 3D models of MDA-MB-468 2015; 26: 487-97.
- Friedrich J, Seidel C, Ebner R, Kunz-Schughart LAJNp. Spheroid-based drug screen: considerations and practical approach. Nat Protoc 2009; 4:309. doi: 10.1038/nprot.2008.226 [Crossref] [ Google Scholar]
- Livak KJ, Schmittgen TDJm. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001; 25:402-8. doi: 10.1006/meth.2001.1262 [Crossref] [ Google Scholar]
- Fomeshi MR, Ebrahimi M, Mowla SJ, Firouzi J, Khosravani PJCJ. CD133 is not suitable marker for isolating melanoma stem cells from D10 cell line. Cell J 2016; 18:21. doi: 10.22074/cellj.2016.3983 [Crossref] [ Google Scholar]
- Roudi R, Madjd Z, Ebrahimi M, Samani FS, Samadikuchaksaraei AJC, letters mb. CD44 and CD24 cannot act as cancer stem cell markers in human lung adenocarcinoma cell line A549. Cell Mol Biol Lett 2014; 19:23. doi: 10.2478/s11658-013-0112-1 [Crossref] [ Google Scholar]
- Segatto I, Baldassarre G, Belletti BJIjoms. STAT3 in breast cancer onset and progression: a matter of time and context. Int J Mol Sci 2018; 19:2818. doi: 10.3390/ijms19092818 [Crossref] [ Google Scholar]
- Galoczova M, Coates P, Vojtesek BJC, letters mb. STAT3, stem cells, cancer stem cells and p63. Cell Mol Biol Lett 2018; 23:12. doi: 10.1186/s11658-018-0078-0 [Crossref] [ Google Scholar]
-
Chung SS, Aroh C, Vadgama JVJPo. Constitutive activation of STAT3 signaling regulates hTERT and promotes stem cell-like traits in human breast cancer cells. PLoS One 2013; 8. 10.1371/journal.pone.0083971.
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MFJPotNAoS. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 2003; 100:3983-8. doi: 10.1073/pnas.0530291100 [Crossref] [ Google Scholar]
- Wu Y, Wu PYJSc, development development. CD133 as a marker for cancer stem cells: progresses and concerns. Stem Cells Dev 2009; 18:1127-34. doi: 10.1089/scd.2008.0338 [Crossref] [ Google Scholar]
-
Sahlberg SH, Spiegelberg D, Glimelius B, Stenerlöw B, Nestor MJPo. Evaluation of cancer stem cell markers CD133, CD44, CD24: association with AKT isoforms and radiation resistance in colon cancer cells. PLoS One 2014; 9. 10.1371/journal.pone.0094621.
- Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J. Molecular definition of breast tumor heterogeneity. Cancer Cell 2007; 11:259-73. doi: 10.1016/j.ccr.2007.01.013 [Crossref] [ Google Scholar]
- Götte M, Yip GWJCr. Heparanase, hyaluronan, and CD44 in cancers: a breast carcinoma perspective. Cancer Res 2006; 66:10233-7. doi: 10.1158/0008-5472.CAN-06-1464 [Crossref] [ Google Scholar]
-
Jaggupilli A, Elkord EJC, Immunology D. Significance of CD44 and CD24 as cancer stem cell markers: an enduring ambiguity. Journal of Immunology Research 2012; 2012. 10.1155/2012/708036.
- Ayob AZ, Ramasamy TSJJobs. Cancer stem cells as key drivers of tumour progression. J Biomed Sci 2018; 25:20. doi: 10.1186/s12929-018-0426-4 [Crossref] [ Google Scholar]
-
Zhou J. MCF7 side population cells and sphere culture as models for breast cancer stem-like cell biology and drug identification. cancerres 2008. 10.1158/0008-5472.CAN-07-2249.
- de la Mare J-A, Sterrenberg JN, Sukhthankar MG, Chiwakata MT, Beukes DR, Blatch GL. Assessment of potential anti-cancer stem cell activity of marine algal compounds using an in vitro mammosphere assay. Cancer Cell Int 2013; 13:39. doi: 10.1186/1475-2867-13-39 [Crossref] [ Google Scholar]
- Wang R, Lv Q, Meng W, Tan Q, Zhang S, Mo X. Comparison of mammosphere formation from breast cancer cell lines and primary breast tumors. J Thorac Dis 2014; 6:829. doi: 10.3978/j.issn.2072-1439.2014.03.38 [Crossref] [ Google Scholar]
- Maiuthed A, Chantarawong W, Chanvorachote PJAr. Lung cancer stem cells and cancer stem cell-targeting natural compounds. Anticancer Res 2018; 38:3797-809. doi: 10.21873/anticanres.12663 [Crossref] [ Google Scholar]
- Wang JY, Chang CC, Chiang CC, Chen WM, Hung SCJJocb. Silibinin suppresses the maintenance of colorectal cancer stem‐like cells by inhibiting PP2A/AKT/mTOR pathways. J Cell Biochem 2012; 113:1733-43. doi: 10.1002/jcb.24043 [Crossref] [ Google Scholar]
- Abdollahi P, Ebrahimi M, Motamed N, Samani FSJA-cd. Silibinin affects tumor cell growth because of reduction of stemness properties and induction of apoptosis in 2D and 3D models of MDA-MB-468. Anticancer Drugs 2015; 26:487-97. [ Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MFJPotNAoS. Prospective identification of tumorigenic breast cancer cells 2003; 100: 3983-8.
- de Beça FF, Caetano P, Gerhard R, Alvarenga CA, Gomes M, Paredes J. Cancer stem cells markers CD44, CD24 and ALDH1 in breast cancer special histological types. J Clin Pathol 2013; 66:187-91. doi: 10.1136/jclinpath-2012-201169 [Crossref] [ Google Scholar]
- Tume L, Paco K, Ubidia-Incio R, Moya JJGMdO. CD133 in breast cancer cells and in breast cancer stem cells as another target for immunotherapy. Gaceta Mexicana de Oncología 2016; 15:22-30. doi: 10.1016/j.gamo.2016.01.003 [Crossref] [ Google Scholar]
- Chang Y-C, Jan C-I, Peng C-Y, Lai Y-C, Hu F-W, Yu C-CJO. Activation of microRNA-494-targeting Bmi1 and ADAM10 by silibinin ablates cancer stemness and predicts favourable prognostic value in head and neck squamous cell carcinomas. Oncotarget 2015; 6:24002. doi: 10.18632/oncotarget.4365 [Crossref] [ Google Scholar]
-
Luo W, Li S, Peng B, Ye Y, Deng X, Yao KJPo. Embryonic stem cells markers SOX2, OCT4 and Nanog expression and their correlations with epithelial-mesenchymal transition in nasopharyngeal carcinoma. PLoS One 2013; 8. 10.1371/journal.pone.0056324.
- Raina K, Kumar S, Dhar D, Agarwal RJJobr. Silibinin and colorectal cancer chemoprevention: a comprehensive review on mechanisms and efficacy. J Biomed Res 2016; 30:452. doi: 10.7555/JBR.30.20150111 [Crossref] [ Google Scholar]
- Weissenstein U, Schumann A, Reif M, Link S, Toffol-Schmidt UD, Heusser PJBc. Detection of circulating tumor cells in blood of metastatic breast cancer patients using a combination of cytokeratin and EpCAM antibodies. BMC Cancer 2012; 12:206. doi: 10.1186/1471-2407-12-206 [Crossref] [ Google Scholar]
- Gusterson BA, Ross DT, Heath VJ, Stein TJBCR. Basal cytokeratins and their relationship to the cellular origin and functional classification of breast cancer. Breast Cancer Res 2005; 7:143. doi: 10.1186/bcr1041 [Crossref] [ Google Scholar]
- Malzahn K, Mitze M, Thoenes M, Moll RJVA. Biological and prognostic significance of stratified epithelial cytokeratins in infiltrating ductal breast carcinomas. Virchows Arch 1998; 433:119-29. doi: 10.1007/s004280050226 [Crossref] [ Google Scholar]
- Ye S, Liu D, Ying Q-LJCoig, development development. Signaling pathways in induced naive pluripotency. Curr Opin Genet Dev 2014; 28:10-5. doi: 10.1016/j.gde.2014.08.002 [Crossref] [ Google Scholar]
- Wei D, Kanai M, Huang S, Xie KJC. Emerging role of KLF4 in human gastrointestinal cancer. Carcinogenesis 2005; 27:23-31. doi: 10.1093/carcin/bgi243 [Crossref] [ Google Scholar]
-
Bai Z-L, Tay V, Guo S-Z, Ren J, Shu M-GJBri. Silibinin induced human glioblastoma cell apoptosis concomitant with autophagy through simultaneous inhibition of mTOR and YAP. Biomed Res Int 2018; 2018. 10.1155/2018/6165192.
- Kim S, Choi MG, Lee HS, Lee SK, Kim SH, Kim WW. Silibinin suppresses TNF-α-induced MMP-9 expression in gastric cancer cells through inhibition of the MAPK pathway. Molecules 2009; 14:4300-11. doi: 10.3390/molecules14114300 [Crossref] [ Google Scholar]